Request Demo
Last update 29 Sep 2025
HIV-1 vaccine(Western University)
Last update 29 Sep 2025
Overview
Basic Info
Drug Type Prophylactic vaccine |
Synonyms Killed-whole HIV-1 vaccine, SAV-001, SAV-001H |
Target- |
Action stimulants |
Mechanism Immunostimulants |
Therapeutic Areas |
Active Indication |
Inactive Indication- |
Originator Organization |
Active Organization |
Inactive Organization- |
License Organization- |
Drug Highest PhasePhase 2 |
First Approval Date- |
Regulation- |
Login to view timeline
Related
2
Clinical Trials associated with HIV-1 vaccine(Western University)NCT01546818
A Phase I, Randomized, Observer-blinded, Placebo-controlled Clinical Study to Assess the Safety, Tolerability, and Immune Response of Killed-Whole HIV-1 Vaccine (SAV001-H) Administered Intramuscularly to Chronic HIV-1 Infected Patients Currently Under Treatment With Highly Active Antiretroviral Therapy (HAART)
The purpose of this study is to examine the safety, tolerability, and immune response to killed-whole HIV-1 (SAV001-H) vaccine as a primary vaccination regimen in HIV infected individuals.
Start Date01 Mar 2012 |
Sponsor / Collaborator |
PER-036-01
Phase II Multicenter Clinical Trial to evaluate the immune response generation capacity and the safety of the ALVAC-HIVVCP1452 Vaccine administered alone and in combination with the MN rpg 120 / HIV-1 vaccine in humans from Brazil, Haiti, Peru and Trinidad and Tobago.
Start Date01 Jan 1900 |
Sponsor / Collaborator |
100 Clinical Results associated with HIV-1 vaccine(Western University)
Login to view more data
100 Translational Medicine associated with HIV-1 vaccine(Western University)
Login to view more data
100 Patents (Medical) associated with HIV-1 vaccine(Western University)
Login to view more data
460
Literatures (Medical) associated with HIV-1 vaccine(Western University)06 Oct 2025·JOURNAL OF EXPERIMENTAL MEDICINE
Design and characterization of HIV-1 vaccine candidates to elicit antibodies targeting multiple epitopes.
Article
Author: Koranda, Nicholas S ; Gnanapragasam, Priyanthi N P ; Nishimura, Yoshiaki ; Nagashima, Kaito ; West, Anthony P ; Segovia, Luisa N ; Hartweger, Harald ; Donau, Olivia K ; Nussenzweig, Michel C ; Bjorkman, Pamela J ; Kakutani, Leesa M ; Keeffe, Jennifer R ; Gavor, Edem ; Martin, Malcolm A ; Gristick, Harry B
A primary goal in the development of an AIDS vaccine is the elicitation of broadly neutralizing antibodies (bNAbs) that protect against diverse HIV-1 strains. To this aim, germline-targeting immunogens have been developed to activate bNAb precursors and initiate the induction of bNAbs. While most preclinical germline-targeting HIV-1 vaccine candidates only include a single bNAb precursor epitope, an effective HIV-1 vaccine will likely require bNAbs that target multiple epitopes on Env. Here, we report a newly designed germline-targeting Env SOSIP trimer, named 3nv.2, that presents three bNAb epitopes on Env: the CD4bs, V3, and V2 epitopes. 3nv.2 forms a stable trimeric Env and binds to bNAb precursors from each of the desired epitopes. Immunization experiments in rhesus macaques and mice demonstrate 3nv.2 elicits the combined effects of its parent immunogens. Our results provide proof of concept for using a germline-targeting immunogen presenting three or more bNAb epitopes and a framework to develop improved next-generation HIV-1 vaccine candidates.
01 Jul 2025·CURRENT HIV RESEARCH
Innovative Single-Cell Sequencing Techniques for B-Cell Analysis and Their Implications for Rational HIV-1 Vaccine Design
Article
Author: Claireaux, Mathieu ; Beaumont, Tim ; van Gils, Marit J. ; Graus, Laura T.M. ; Guerra, Denise
Abstract::
The application of single-cell analysis to investigate immune cell diversity has historically
been considered a complex task. Recently, innovative techniques have emerged revolutionizing
the way immune cells can be explored, offering unprecedented insights into the dynamics of
this complex system. In particular, novel approaches have enabled a detailed characterization of
B-cell responses, encompassing immune repertoire, gene expression, and phenotype analysis at an
individual cell level. By analyzing single B-cells, researchers can unravel their heterogeneity,
trace clonal evolution, and track immune responses during infections and vaccinations over time,
gaining a deeper understanding of the mechanisms underlying antibody secretion and immune memory
formation. This knowledge can inform the development of optimal immunogens, adjuvants,
and vaccine platforms, which are crucial for inducing robust, long-lasting protective responses
and overcoming existing challenges in vaccine research. This is particularly valuable for rational
vaccine design against certain pathogens, such as human immunodeficiency virus [HIV-1], for
which a successful vaccine remains to be developed due to the need to elicit rare broadly neutralizing
antibodies that target conserved epitopes on the genetically diverse envelope glycoprotein
trimer. This review will highlight the latest advancements in single-cell sequencing techniques
and bioinformatic tools for the analysis of B-cell responses in the context of infectious diseases
and vaccinations. Single-cell sequencing techniques, their applications, and their pivotal role in advancing
the design of next-generation vaccines, especially in the context of HIV-1, will be discussed.
10 Jun 2025·CURRENT HIV RESEARCH
Germline-targeting Strategies to Induce bNAbs against HIV-1
Article
Author: Aldon, Yoann ; Atabey, Tugba ; Sanders, Rogier W
Abstract::
Developing an effective HIV-1 vaccine remains a critical global health challenge, hin-dered by the virus’s high genetic diversity, immune evasion strategies, and structural complexity of its Envelope (Env) glycoprotein. Broadly neutralizing antibodies (bNAbs), capable of targeting conserved Env epitopes, offer a promising path for vaccine design. Germline-targeting (GT) strat-egies have emerged as a promising approach to engage naive B cell precursors that have the po-tential to mature into bNAb-producing cells. Advances in GT have enabled the design of immu-nogens capable of recruiting specific bNAb precursors in animal models and early clinical trials. Despite these successes, achieving neutralization breadth requires sequential immunizations with tailored boosting strategies to guide B cell maturation. Studies underscore the importance of using immunogens that mimic native Env structures while modulating glycosylation patterns to focus immune responses. Emerging approaches, such as membrane-bound presentation and mRNA de-livery, hold the potential for enhancing immunogen effectiveness and rapid pre-clinical and in human screening to identify combinations of immunogens that foster bNAb lineages. This review seeks to synthesize key developments in GT strategies for HIV-1 vaccines, highlighting the de-sign and implementation of immunogens that drive bNAb precursor maturation. It aims to under-score the importance of integrating structural insights, immunogen sequence design, and delivery methods to enhance the induction of bNAbs, offering direction for future research to address ex-isting gaps and optimize vaccine efficacy.
2
News (Medical) associated with HIV-1 vaccine(Western University)29 Oct 2024
NANTES, France--(
BUSINESS WIRE
)--Naobios, a CDMO (Contract Development and Manufacturing Organization) providing bioprocess development and GMP production of clinical batches of virus-based products, and Sumagen Canada Inc (Sumagen), a Korean-Canadian biotechnology company part of CreoSG Co., Ltd, today announce the production of HIV-1 vaccine candidate at bench scale.
This key milestone, achieved on schedule, will allow Naobios to scale up the production of Sumagen’s HIV-1 vaccine followed by cGMP production to launch phase I/II clinical trials by the end of 2025.
“We are thrilled to have reached such a strategic industrial milestone within expected initial timelines, which is extremely significant due to the initial project delays resulting from the Covid-19 pandemic. This achievement solidifies our trust in Naobios to help our HIV-1 vaccine reach the crucial phase II trials, bringing us closer to delivering a vaccine to patients in need,” said Dr Sangkyun Lee, president of Sumagen.
“To have reached the process development and optimization stage within the challenging initial planned timelines speaks volumes of our capabilities and decades of experience in viral process development. We are proud to be working with innovators like Sumagen who have the ability to significantly impact global human health,” said Eric Le Forestier, general manager of Naobios.
Sumagen candidate (SAV001) is a genetically modified, whole-killed HIV vaccine which
demonstrated both tolerance and safety for human use
in phase I trials.
About Naobios
Naobios is a Contract Development and Manufacturing Organization (CDMO) providing bioprocess development and offering GMP production of clinical batches of BSL2/BSL3 viral vaccines, oncolytic viruses, viral vectors and challenge agents. Naobios joined the Clean Biologics group in 2019.
Having built up 15 years’ experience in bioprocess development, Naobios helps its clients to bring their drug candidates to the clinical stage as rapidly as possible – at the highest level of quality – whilst building on its technical know-how in scalable and industrial processes. With its adaptability and range of skills, the company can lead a project from the initial stages through to completion, with a motivated and dedicated team. Its highly qualified staff have the experience to deal with a wide range of viruses, as well as multiple cell substrate lines. It currently has 40 staff.
www.naobios.com
Vaccine
14 Nov 2023
Longevity of neutralizing antibodies is an essential factor for an effective HIV-1 vaccination.
An international team has for the first time researched the longevity of neutralizing antibodies in HIV-1-infected people. Currently, it is assumed that an HIV-1 vaccine can only be effective if it produces these antibodies in vaccinated humans. The findings improve understanding of the dynamics of such antibodies and are an important building block for further research into an HIV-1 vaccine. Professor Dr Florian Klein, Director of the Institute of Virology at the University Hospital Cologne, and Dr Dr Philipp Schommers, Head of the Laboratory for Antiviral Immunity at Department I of Internal Medicine of the University Hospital Cologne, were involved in the study. The publication was published under the title 'Dynamics and durability of HIV-1 neutralization are determined by viral replication' in the journal Nature Medicine.
Dr Dr Schommers, first author of the study, reports: "We were able to show that the HIV-1 neutralization activity in patients strongly depends on the amount of virus in patients. While this dependence could be investigated in other infectious diseases, such as COVID-19, shortly after the initial description of the disease, the longevity of neutralizing antibodies in HIV-1 had not yet been shown in large studies."
Despite effective drugs that are the basis for treating the HIV-1 infection and can effectively prevent the transmission of the virus, over 1.2 million people are infected with HIV every year. Therefore, the development of an effective HIV-1 vaccine is still being intensely researched. So-called broadly neutralizing antibodies (bNAbs) can prevent HIV-1 infection. Researchers are trying to induce such bNAbs through a vaccination in humans. However, this has proven to be extremely difficult. Therefore, vaccines that allow the formation of bNAbs in humans have not yet been developed. It is also unclear how long such broadly neutralizing antibodies remain in humans. However, this knowledge is extremely important in developing successful strategies for an HIV-1 vaccination.
The researchers led by Professor Klein and Dr Schommers have therefore investigated the HIV-1 antibody response in more than 2,300 patients from Germany, Tanzania, Cameroon and Nepal. They were able to identify various factors that cause patients to form neutralizing antibodies naturally. In addition, they identified so-called 'elite neutralizers', i.e. HIV-1-infected individuals who build up a very potent and broadly neutralizing antibody response. When the international research team examined HIV-1-infected individuals over time, it was then able to find out with what dynamics HIV-1 neutralizing antibodies can be sustained or the concentration of these antibodies can be further decreased in the blood. Here it was shown that the antibody response in these patients decreases over the years, but in them highly potent bNAbs are detectable even after years. This is an important finding and indicates that a possible HIV-1 vaccine can trigger a permanent vaccine response.
VaccineImmunotherapyClinical Study
100 Deals associated with HIV-1 vaccine(Western University)
Login to view more data
R&D Status
10 top R&D records. to view more data
Login
| Indication | Highest Phase | Country/Location | Organization | Date |
|---|---|---|---|---|
| HIV Infections | Phase 2 | United States | - |
Login to view more data
Clinical Result
Clinical Result
Indication
Phase
Evaluation
View All Results
| Study | Phase | Population | Analyzed Enrollment | Group | Results | Evaluation | Publication Date |
|---|
No Data | |||||||
Login to view more data
Translational Medicine
Boost your research with our translational medicine data.
login
or

Deal
Boost your decision using our deal data.
login
or

Core Patent
Boost your research with our Core Patent data.
login
or
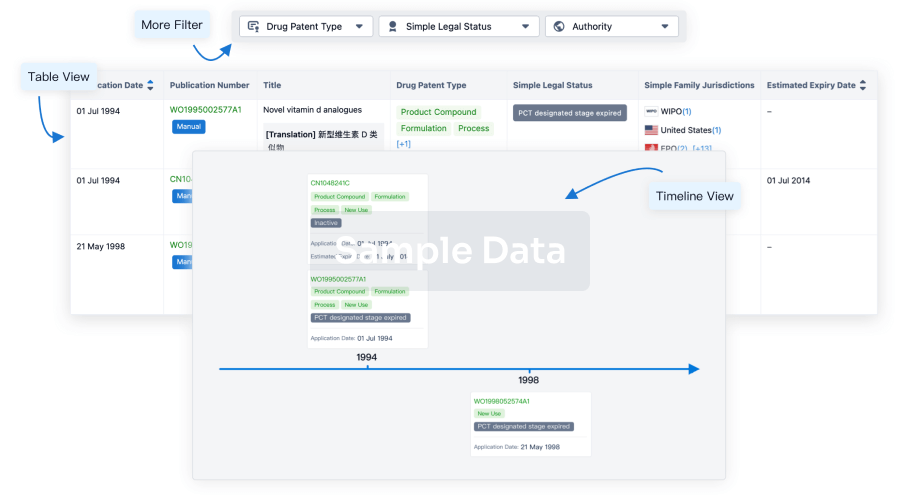
Clinical Trial
Identify the latest clinical trials across global registries.
login
or

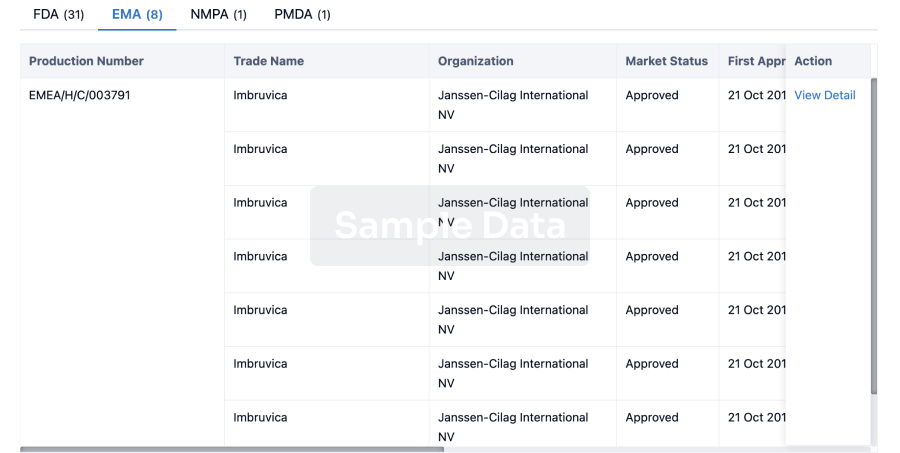
Approval
Accelerate your research with the latest regulatory approval information.
login
or
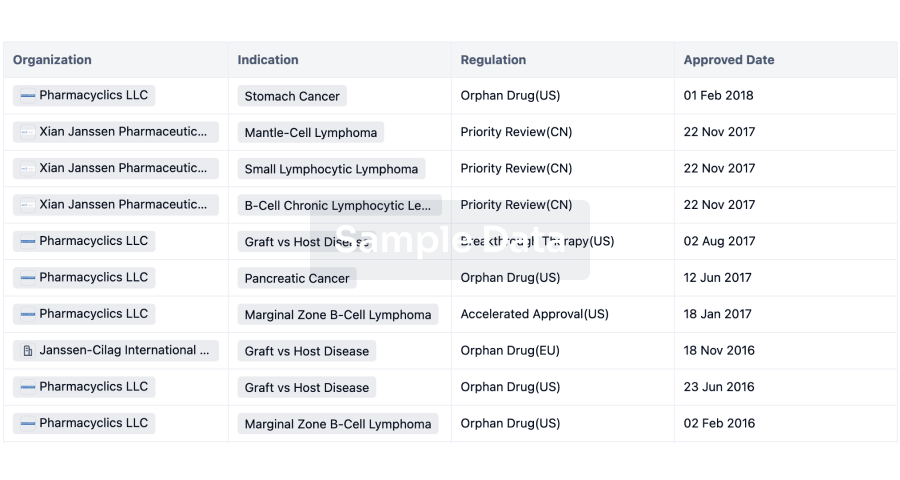
Regulation
Understand key drug designations in just a few clicks with Synapse.
login
or
AI Agents Built for Biopharma Breakthroughs
Accelerate discovery. Empower decisions. Transform outcomes.
Get started for free today!
Accelerate Strategic R&D decision making with Synapse, PatSnap’s AI-powered Connected Innovation Intelligence Platform Built for Life Sciences Professionals.
Start your data trial now!
Synapse data is also accessible to external entities via APIs or data packages. Empower better decisions with the latest in pharmaceutical intelligence.
Bio
Bio Sequences Search & Analysis
Sign up for free
Chemical
Chemical Structures Search & Analysis
Sign up for free
