Request Demo
Last update 07 Feb 2026
SMARCA2 inhibitors (Arvinas LLC)
Last update 07 Feb 2026
Overview
Basic Info
Originator Organization- |
Active Organization |
Inactive Organization- |
License Organization- |
Drug Highest PhasePreclinical |
First Approval Date- |
Regulation- |
Related
100 Clinical Results associated with SMARCA2 inhibitors (Arvinas LLC)
Login to view more data
100 Translational Medicine associated with SMARCA2 inhibitors (Arvinas LLC)
Login to view more data
100 Patents (Medical) associated with SMARCA2 inhibitors (Arvinas LLC)
Login to view more data
4
Literatures (Medical) associated with SMARCA2 inhibitors (Arvinas LLC)13 Mar 2025·ACS Medicinal Chemistry Letters
Novel 1,6-Naphthridine Compounds as SMARCA2 Inhibitors for Treating Non-small Cell Lung Cancer
Author: Sabnis, Ram W.
Provided herein are novel 1,6-naphthridine compounds as SMARCA2 inhibitors, pharmaceutical compositions, use of such compounds in treating non-small cell lung cancer and processes for preparing such compounds.
01 Mar 2025·EUROPEAN JOURNAL OF MEDICINAL CHEMISTRY
SMARCA2 protein: Structure, function and perspectives of drug design
Review
Author: Yang, Xinyu ; Guo, Zhaolin ; Cao, Shuang ; Jiang, Sisi ; Han, Yuxuan ; Wang, Peng
SMARCA2 is an ATPase that regulates chromatin structure via ATP pathways, controlling cell division and differentiation. SMARCA2's bromodomain and ATPase domain, crucial for chromatin remodeling and cell regulation, are therapeutic targets in cancer treatment. This review explores the role of SMARCA2 in cancer development by studying its protein structure and physiological functions. It further discusses the roles and distinctions of SMARCA2 and its related family proteins in cancer. Additionally, this article categorizes known SMARCA2 inhibitors into four classes based on their basic structure and examines their structure-activity relationships (SAR). This review outlines the structural mechanisms of SMARCA2 inhibitors, highlighting interactions with specific amino acids. By analyzing the SAR of inhibitors, we propose a tailored inhibitor model for the bromodomain of SMARCA2, emphasizing α, γ-H-bond donors/acceptors, and β-rigid structures as crucial for effective binding. This research provides guidance for the design and optimization of future drugs targeting the SMARCA2 protein.
25 Jan 2024·Journal of medicinal chemistryQ1 · MEDICINE
PROTACs Targeting BRM (SMARCA2) Afford Selective In Vivo Degradation over BRG1 (SMARCA4) and Are Active in BRG1 Mutant Xenograft Tumor Models
Q1 · MEDICINE
Article
Author: Hole, Alison J. ; Ye, Xiaofen ; Ye, Crystal S. ; Quinn, Connor ; Berlin, Michael ; Chen, Huifen ; Harbin, Alicia ; Li, Limei ; Cantley, Jennifer ; Chan, Emily W. ; Pizzano, Jennifer ; Wang, Jing ; Soto, Leofal ; Davenport, Kim ; DiNicola, Dean ; Gordon, Debbie ; Koenig, Stefan G. ; Pérez-Dorado, Inmaculada ; Cheng, Yunxing ; Hamman, Brian D. ; He, Mingtao ; Januario, Thomas ; Tian, Qingping ; Dragovich, Peter S. ; Bookbinder, Mark ; Sun, Hongming ; Kerry, Philip S. ; Wang, Weifeng ; Staben, Leanna R. ; Zhang, Penghong ; Cheung, Tommy K. ; Yauch, Robert ; Bortolon, Elizabeth ; Chen, Xin ; Rose, Christopher M. ; Broccatelli, Fabio ; Merchant, Mark ; Cadelina, Greg ; Haskell, Roy ; Rousseau, Emma ; Zhou, Yuhui
The identification of VHL-binding proteolysis targeting chimeras (PROTACs) that potently degrade the BRM protein (also known as SMARCA2) in SW1573 cell-based experiments is described. These molecules exhibit between 10- and 100-fold degradation selectivity for BRM over the closely related paralog protein BRG1 (SMARCA4). They also selectively impair the proliferation of the H1944 "BRG1-mutant" NSCLC cell line, which lacks functional BRG1 protein and is thus highly dependent on BRM for growth, relative to the wild-type Calu6 line. In vivo experiments performed with a subset of compounds identified PROTACs that potently and selectively degraded BRM in the Calu6 and/or the HCC2302 BRG1 mutant NSCLC xenograft models and also afforded antitumor efficacy in the latter system. Subsequent PK/PD analysis established a need to achieve strong BRM degradation (>95%) in order to trigger meaningful antitumor activity in vivo. Intratumor quantitation of mRNA associated with two genes whose transcription was controlled by BRM (PLAU and KRT80) also supported this conclusion.
3
News (Medical) associated with SMARCA2 inhibitors (Arvinas LLC)22 Oct 2025
Three posters to be presented featuring pre-clinical data of potential best-in-class FGFR3 selective inhibitor, SMARCA2 selective inhibitor, and P53 Y220C reactivatorCandidates for FGFR3 and SMARCA2 programs identified: IND enabling studies anticipated in mid-2026 Leuven, Belgium, October 22, 2025. Onco3R Therapeutics, a clinical-stage immunology and oncology biotech company dedicated to transforming patients’ lives with best-in-class medicines, today announced that it will present preclinical data from its FGFR3, SMARCA2 and P53 Y220C small molecules programs in 3 posters at the EORTC-NCI-AACR Symposium on Molecular Targets and Cancer Therapeutics, taking place October 22-26, 2025, in Boston. “First generation precision medicines are often suboptimal in the clinic due to low target coverage, off-target toxicity and emergence of resistance. At Onco3R, our vision is to design best-in-class medicines to address the unmet needs left by first generation drugs and unlock the full potential of therapeutic targets” said François Gonzalvez, PhD, CSO and co-Founder of Onco3R Therapeutics. “We are thrilled to present the first preclinical data from our lead oncology programs FGFR3, SMARCA2 and P53 Y220C. Each program has identified best-in-class molecules which offer the potential to deliver transformational efficacy and improved tolerability for patients. Our FGFR3 and SMARCA2 candidates, G-012 and G-141 respectively, have reached the optimum potency and selectivity profile to mitigate dose-limiting toxicities while maintaining maximum target coverage. This has translated into robust anti-tumor activity in vivo. The poster presentations will highlight data supporting the advancement of these two candidates towards the clinic, as well as the discovery of unique small molecule P53 reactivators.” “These compelling preclinical results further validate our patient-centric drug discovery approach, which integrates deep translational science with rational, structure-based and AI-augmented drug design”, Pierre Raboisson, PhD, CEO and co-Founder of Onco3R Therapeutics said. “We look forward to advancing these two candidates and remain on track to initiate IND-enabling studies in mid-2026. The identification of these candidates, alongside the continued clinical progress of our SIK3 inhibitor O3R-5671 in autoimmune indications, reinforces Onco3R’s strong strategic position. With a robust pipeline and clear execution momentum, we are confidently advancing toward our next value-driving milestones.” Presentation details Title: Discovery of Best-in-Class FGFR3 small molecule inhibitors with high isoform selectivity and activity against gatekeeper mutationsSession:
Session: Poster Session C
Session Date and Time: Saturday, October 25, 12:30-4pmPresenting author: Sandrine Vendeville, PhD Key findings from preclinical studies include: G-012 demonstrated best-in-class potency and selectivity with favorable drug-like properties.Based on translational modelling, the compound reached the optimal selectivity against other FGFR isoforms to mitigate off-target toxicity and maintain maximal target coverage.G-012 showed robust anti-proliferative activity in FGFR3-driven cancer cells and induced significant tumor regression in vivo.G-012 is currently advancing in 14 days toxicology studies.IND-enabling studies are anticipated in mid-2026. Title: Discovery of novel SMARCA2 small molecule inhibitors with best-in-class potency and selectivity for the treatment of SMARCA4-mutant cancersSession: Poster Session C
Session Date and Time: Saturday, October 25, 12:30-4pmPresenting author: Lijs Beke, PhD Key findings from preclinical studies include: G-141 combined best-in-class cellular potency and selectivity to allow optimal target coverage and unlock the full therapeutic potential of SMARCA2 inhibition.The compound showed synthetic lethality in SMARCA4-deficient cells and induced robust anti-tumor activity in vivo without signs of SMARCA4-related toxicity.G-141 showed favorable drug-like properties and is currently advancing in 14 days toxicology studies.IND-enabling studies are anticipated in mid-2026. Title: Discovery of a Best-in-Class small molecule p53 Y220C reactivator: Breaking through the potency ceilingSession: Poster Session C
Session Date and Time: Saturday, October 25, 12:30-4pmPresenting author: François Gonzalvez, PhD Key findings from preclinical studies include: Onco3R patient-centric drug discovery approach identified unique small molecule P53 reactivators with best-in-class cellular potency.Onco3R leads exhibit the optimal potency and residence time to induce deep and sustain target engagement and fully unlock the tumor suppressive function of P53 in cells. This translated into robust anti-proliferative activity in P53 Y220C mutant cancer cell lines (single digit nanomolar IC50s) and tumor regression in a Y220C P53 mutant xenograft model.Further characterization of the lead candidates is ongoing. About Onco3R TherapeuticsAt Onco3R Therapeutics, we are driven by our purpose to transform the lives of patients with autoimmune diseases and cancer through precision-designed, best-in-class therapies. With over 150 years of combined R&D experience, our team brings deep expertise in disease biology, drug discovery & development, and translational science. We focus on clinically validated targets and select the right therapeutic modality, small or large molecules, to address the underlying disease biology with best-in-class therapies. Our mission is to develop safer, more effective medicines in oncology and immunology that truly make a difference for patients. By integrating learnings from past clinical challenges and applying cutting-edge technologies, we aim to de-risk clinical development and accelerate the delivery of innovative treatments with real-world impact. The company is based in the biotech cluster in Leuven, Belgium. For more information, visit www.onco3r.com or follow us on LinkedIn. About O3R-5671O3R-5671 has been developed based on more than 12 years of preclinical and clinical data on SIK inhibitors for autoimmune diseases. O3R-5671 is a highly selective SIK3 inhibitor, designed to avoid the toxicities associated with inhibiting SIK1 and SIK2. Furthermore, O3R-5671 does not inhibit other kinases and has demonstrated a highly attractive profile in an extensive safety panel. Preclinical data demonstrated that O3R-5671 inhibits the release of the inflammatory cytokines TNFα and IL-23 and promotes the release of the immunomodulatory cytokine IL-10. These data, along with data from animal models of autoimmune diseases, indicate that O3R-5671 has the potential to treat a variety of autoimmune diseases including ulcerative colitis, Crohn’s Disease, psoriasis, psoriatic arthritis and rheumatoid arthritis. O3R-5671 is currently being investigated in a first-in-human study in healthy volunteers with a SAD-MAD design.
Attachment
AACR-EORTC-NCI Meeting 2025_final
AACRImmunotherapyClinical Result
05 Aug 2025
FHD-909 (LY4050784) Phase 1 dose escalation trial in SMARCA4 (BRG1) mutated cancers, with non-small cell lung cancer (NSCLC) as the primary target population, is enrolling well and remains on track
Preclinical synergistic benefit of FHD-909 in combination with pembrolizumab and KRAS inhibitors reinforces significant potential in NSCLC Selective CBP degrader on track for IND-enabling studies, targeting an IND in 2026 Continued progress on Selective EP300 degrader and Selective ARID1B degrader, with program updates expected in Q4 2025 Strong balance sheet with cash, cash equivalents, and marketable securities of $198.7 million as of June 30, 2025; cash runway into 2028 CAMBRIDGE, Mass., Aug. 05, 2025 (GLOBE NEWSWIRE) -- Foghorn® Therapeutics Inc. (Nasdaq: FHTX), a clinical-stage biotechnology company pioneering a new class of medicines that treat serious diseases by correcting abnormal gene expression, today provided a financial and corporate update in conjunction with the Company’s 10-Q filing for the quarter ended June 30, 2025. With an initial focus in oncology, Foghorn’s Gene Traffic Control® Platform and resulting broad pipeline have the potential to transform the lives of people suffering from a wide spectrum of diseases. “We continue to make meaningful progress advancing our pipeline to treat a wide range of cancers,” said Adrian Gottschalk, President and Chief Executive Officer of Foghorn. “The FHD-909 dose escalation trial, which is part of our strategic collaboration with Lilly, is enrolling well and remains on track. Additionally, preclinical synergistic activity of FHD-909 in combination with KRAS inhibitors and pembrolizumab supports clinical exploration of FHD-909 in difficult-to-treat NSCLC.” Mr. Gottschalk continued, “Our wholly owned selective degrader programs targeting CBP, EP300 and ARID1B, continue to advance with strong momentum. Our Selective CBP degrader has shown encouraging activity in ER+ breast cancer with potential beyond EP300-mutant tumors, and we are targeting an IND in 2026. Additionally, we anticipate program updates in the fourth quarter of 2025 for both our Selective EP300 degrader, which has shown robust anti-tumor activity across a range of hematologic malignancies, and our Selective ARID1B degrader, which has achieved selective targeted degradation. Backed by a strong balance sheet and a cash runway into 2028, we are well positioned to further advance our differentiated programs.” Program Overview and Upcoming Milestones FHD-909 (LY4050784). FHD-909 is a first-in-class oral SMARCA2 selective inhibitor that has demonstrated in preclinical studies to have high selectivity over its closely related paralog SMARCA4, two proteins that are the catalytic engines across all forms of the BAF complex. Selectively blocking SMARCA2 activity is a promising synthetic lethal strategy intended to induce tumor death while sparing healthy cells. SMARCA4 is mutated in up to 10% of NSCLC alone and implicated in a significant number of solid tumors. Phase 1 trial enrolling well and remains on track. Enrollment in the first-in-human Phase 1 multi-center trial of FHD-909 is progressing well. The trial in patients with NSCLC as the primary target population is on track, following the dosing of the first patient in October 2024. At the American Association for Cancer Research (AACR) Annual Meeting in April 2025, Lilly presented, on behalf of the collaboration, the clinical study design poster for the Phase 1 trial evaluating FHD-909 in patients with SMARCA4 mutated locally advanced or metastatic solid tumors who have exhausted standard treatment options. The primary target population is NSCLC. Synergistic preclinical data of FHD-909 in combination with pembrolizumab and KRAS inhibitors. Preclinical data presented at AACR demonstrates enhanced anti-tumor activity of FHD-909 in combination with standard-of-care (SoC) chemotherapies, anti-PD-1 pembrolizumab and several novel KRAS inhibitors in NSCLC animal models. The combination data will inform further development plans of FHD-909. Ongoing strategic collaboration with Lilly. Foghorn is collaborating with Lilly to develop novel oncology medicines, including a U.S. 50/50 U.S. co-development and co-commercialization agreement for its selective SMARCA2 oncology program that includes both a selective inhibitor and a selective degrader, as well as an additional undisclosed oncology target. The collaboration also includes three discovery programs from Foghorn’s proprietary Gene Traffic Control® platform. Selective CBP degrader program. Foghorn's Selective CBP degrader selectively targets CBP in EP300-mutated cancer cells, which are found in various cancer types, including bladder, gastric, and endometrial cancers. CBP and EP300 are closely related acetyltransferases, and when EP300 is mutated, this creates a synthetic lethal relationship that the therapy exploits. Attempts to selectively drug CBP have been challenging due to the high level of similarity between the two proteins, while dual inhibition of CBP/EP300 has been associated with dose-limiting toxicities. Presented preclinical combination data with Selective CBP degrader in ER+ breast cancer. In April 2025, preclinical data showing Selective CBP degraders have combination benefit with approved chemotherapies and targeted agents in solid tumors beyond EP300-mutant cancers was presented as a poster at the AACR Annual Meeting. Synergistic combination activity demonstrated including with paclitaxel and CDK4/6 inhibitor abemaciclib in ER+ breast cancer.Findings support combination opportunities for selective CBP degraders in solid tumors beyond EP300-mutant cancers. On track for IND-enabling studies, targeting an IND in 2026. Selective EP300 degrader program. Foghorn is developing a Selective EP300 degrader for the treatment of hematological malignancies and prostate cancer. Attempts to selectively drug EP300 have been challenging due to the high level of similarity between EP300 and CBP, while dual inhibition of CBP/EP300 has been associated with dose limiting toxicities. EP300 lineage dependencies are established in diffuse large b-cell lymphoma (DLBCL) and multiple myeloma (MM). Presented preclinical data showing combination with DLBCL and MM SoC in April 2025. Anti-tumor activity in a broad range of hematological malignancies in vitro, including DLBCL, MM, and follicular lymphoma.Combination of EP300 degrader with SoC in both DLBCL and MM are highly synergistic in vitro.Selective EP300 degradation is effective in IMiD-resistant MM cell lines. Program update expected in Q4 2025. Selective ARID1B degrader program. Foghorn's Selective ARID1B degrader selectively targets and degrades ARID1B in ARID1A-mutated cancers. ARID1A is the most mutated subunit in the BAF complex and amongst the most mutated proteins in cancer. These mutations lead to a dependency on ARID1B in several types of cancer, including ovarian, endometrial, colorectal, and bladder. Attempts to selectively drug ARID1B have been challenging because of the high degree of similarity between ARID1A and ARID1B and the fact that ARID1B has no enzymatic activity to target. ARID1B is a major synthetic lethal target implicated in up to 5% of all solid tumors. Developed highly potent and selective binders. Preclinical data demonstrated potent and selective small molecule binders to ARID1B.Selective degradation of ARID1B achieved. Foghorn has successfully selectively degraded ARID1B.Program update expected in Q4 2025. Chromatin Biology and Degrader Platform. Foghorn continues to advance its chromatin biology and degrader platform with investments in novel ligases, long-acting injectables, oral delivery, and induced proximity. Second Quarter 2025 Financial Highlights Collaboration Revenue. Collaboration revenue was $7.6 million for the three months ended June 30, 2025, compared to $6.9 million for the three months ended June 30, 2024. The increase was driven by the continued advancement of programs under the Lilly Collaboration Agreement.Research and Development Expenses. Research and development expenses were $21.8 million for the three months ended June 30, 2025, compared to $23.8 million for the three months ended June 30, 2024. The decrease is attributed to a decrease in FHD-286 costs, decreases in personnel-related costs, early development and other research external costs and facilities and IT-related expenses, partially offset by an increase in Lilly-partnered programs.General and Administrative Expenses. General and administrative expenses were $6.9 million for the three months ended June 30, 2025, compared to $7.3 million for the three months ended June 30, 2024. This decrease was primarily due to lower consulting costs and lower facilities and IT related expenses.Net Loss. Net loss was $17.9 million for the three months ended June 30, 2025, compared to a net loss of $23.0 million for the three months ended June 30, 2024.Cash, Cash Equivalents, and Marketable Securities. As of June 30, 2025, the Company had $198.7 million in cash, cash equivalents, and marketable securities, providing cash runway into 2028. About FHD-909FHD-909 (LY4050784) is a potent, first-in-class, allosteric, and orally available small molecule that selectively inhibits the ATPase activity of SMARCA2 (BRM) over its closely related paralog SMARCA4 (BRG1), two proteins that are the catalytic engines across all forms of the BAF complex, one of the key regulators of the chromatin regulatory system. In preclinical studies, tumors with mutations in SMARCA4 rely on SMARCA2 for their survival. FHD-909 has shown significant anti-tumor activity across multiple SMARCA4-mutant lung tumor models. About Foghorn TherapeuticsFoghorn® Therapeutics is discovering and developing a novel class of medicines targeting genetically determined dependencies within the chromatin regulatory system. Through its proprietary scalable Gene Traffic Control® platform, Foghorn is systematically studying, identifying, and validating potential drug targets within the chromatin regulatory system. The Company is developing multiple product candidates in oncology. Visit our website at www.foghorntx.com for more information on the Company, and follow us on X and LinkedIn. Forward-Looking StatementsThis press release contains “forward-looking statements.” Forward-looking statements include statements regarding the Company’s ongoing Phase 1 trial of FHD-909 in SMARCA4-mutated cancers, pre-clinical product candidates, expected timing of clinical data, expected cash runway, expected timing of regulatory filings, and research efforts and other statements identified by words such as “could,” “may,” “might,” “will,” “likely,” “anticipates,” “intends,” “plans,” “seeks,” “believes,” “estimates,” “expects,” “continues,” “projects” and similar references to future periods. Forward-looking statements are based on our current expectations and assumptions regarding capital market conditions, our business, the economy and other future conditions. Because forward-looking statements relate to the future, by their nature, they are subject to inherent uncertainties, risks and changes in circumstances that are difficult to predict. As a result, actual results may differ materially from those contemplated by the forward-looking statements. Important factors that could cause actual results to differ materially from those in the forward-looking statements include regional, national or global political, economic, business, competitive, market and regulatory conditions, including risks relating to our clinical trials and other factors set forth under the heading “Risk Factors” in the Company’s Annual Report on Form 10-K for the year ended December 31, 2024, as filed with the Securities and Exchange Commission. Any forward-looking statement made in this press release speaks only as of the date on which it is made.
Condensed Consolidated Balance Sheets(In thousands) June 30, 2025 December 31, 2024Cash, cash equivalents and marketable securities$198,665 $243,747 All other assets 27,571 40,235 Total assets$226,236 $283,982 Deferred revenue, total$266,554 $280,063 All other liabilities 36,341 49,447 Total liabilities$302,895 $329,510 Total stockholders’ deficit$(76,659) $(45,528)Total liabilities and stockholders’ deficit$226,236 $283,982
Condensed Consolidated Statements of Operations(In thousands, except share and per share amounts) Three Months Ended June 30, 2025 2024 Collaboration revenue$7,557 $6,888 Operating expenses: Research and development 21,792 23,797 General and administrative 6,862 7,325 Impairment of long-lived assets — 2,398 Total operating expenses$28,654 $33,520 Loss from operations$(21,097) $(26,632)Total other income, net$3,161 $3,653 Net loss$(17,936) $(22,979)Net loss per share attributable to common stockholders—basic and diluted (0.28) (0.45)Weighted average common shares outstanding—basic and diluted 62,978,219 51,580,310
Contact:Karin Hellsvik, Foghorn Therapeutics Inc.khellsvik@foghorntx.com
Phase 1Financial StatementAACR
14 May 2025
FHD-909 (LY4050784) advancing in Phase 1 dose escalation trial in SMARCA4 (BRG1) mutated cancers, with non-small cell lung cancer (NSCLC) as the primary target population Data presented at AACR show synergistic activity with FHD-909 in combination with pembrolizumab and KRAS inhibitors and support clinical exploration Selective CBP degrader on track for IND-enabling studies, targeting IND in 2026 Continued progress on Selective EP300 degrader and Selective ARID1B degrader with program updates expected in H2 2025 Strong balance sheet with cash, cash equivalents, and marketable securities of $220.6 million as of March 31, 2025, provides cash runway into 2027 CAMBRIDGE, Mass., May 14, 2025 (GLOBE NEWSWIRE) -- Foghorn® Therapeutics Inc. (Nasdaq: FHTX), a clinical-stage biotechnology company pioneering a new class of medicines that treat serious diseases by correcting abnormal gene expression, today provided a financial and corporate update in conjunction with the Company’s 10-Q filing for the quarter ended March 31, 2025. With an initial focus in oncology, Foghorn’s Gene Traffic Control® Platform and resulting broad pipeline have the potential to transform the lives of people suffering from a wide spectrum of diseases. “In the first quarter, we continued to progress our novel partnered and proprietary programs, and we were pleased to share more details on the pipeline progress at this year’s AACR annual meeting. The FHD-909 Phase 1 dose escalation trial is proceeding apace. Importantly, the FHD-909 combination data with chemotherapies, KRAS inhibitors and pembrolizumab, that was shared at AACR, reinforces the expansive potential of the selective SMARCA2 inhibitor program in non-small cell lung cancer,” said Adrian Gottschalk, President and Chief Executive Officer of Foghorn. “Initial data defining potential combination opportunities in ER+ breast cancer for our Selective CBP degrader program are encouraging and support potential beyond EP300-mutant cancers. Our Selective EP300 degrader program continues to show anti-proliferative activity in a broad range of hematological malignancies. And lastly, as previously reported, we have achieved selective degradation for our Selective ARID1B program and look forward to providing a program update in 2025.” Mr. Gottschalk continued, “These advancements with FHD-909 and our pipeline programs underscore our track record and leadership in engineering promising selective therapeutics. With our robust balance sheet and cash runway into 2027, we are in a strong position to deliver significant value with differentiated, high-impact therapeutics in 2025 and beyond.” Recent Corporate Updates Data presented at American Association for Cancer Research (AACR) Annual Meeting. In April 2025, Foghorn presented new preclinical data highlighting pipeline progress for potential first-in-class medicines including FHD-909, a SMARCA2 (BRM) selective inhibitor, and the Selective CBP degrader program and Selective EP300 degrader program. Two Appointments to Board of Directors. In May, Foghorn announced the appointments of Neil Gallagher, M.D., Ph.D. and Stuart Duty, to its Board of Directors. Dr. Gallagher brings over 20 years of executive experience at pharmaceutical and biopharmaceutical organizations leading drug development programs across several therapeutic areas, including oncology. Currently he serves as the President, Head of Research and Development at Syndax Pharmaceuticals and held prior clinical development leadership roles at AbbVie, Amgen, Novartis, and AstraZeneca.Mr. Duty has over 30 years of executive experience in finance and investment banking in biotechnology and specialty pharmaceuticals. Most recently, he served as a Senior Managing Director at Guggenheim Securities, LLC where he advised senior executives and boards on a range of financing activities and strategic transactions. Previously, he held senior roles at Piper Jaffray and Montgomery Securities and operating roles at Oracle Partners and Curative Technologies. Second Annual Chromatin Regulation Summit. In May 2025, Foghorn hosted its “Chromatin Regulation Summit: Targeted Protein Degradation and Induced Proximity,” at their corporate headquarters in Cambridge, Massachusetts. The live event featured presentations and panel discussions with world-renowned industry and academic key opinion leaders on current and future applications of targeted protein degradation and induced proximity modalities for the treatment of disease. Program Overview and Upcoming Milestones FHD-909 (LY4050784). FHD-909 is a first-in-class oral SMARCA2 selective inhibitor that has demonstrated in preclinical studies to have high selectivity over its closely related paralog SMARCA4, two proteins that are the catalytic engines across all forms of the BAF complex. Selectively blocking SMARCA2 activity is a promising synthetic lethal strategy intended to induce tumor death while sparing healthy cells. SMARCA4 is mutated in up to 10% of NSCLC alone and implicated in a significant number of solid tumors. Advancing Phase 1 trial. Ongoing enrollment and dose escalation in the first-in-human Phase 1, multicenter trial for FHD-909 in SMARCA4 mutated cancers, with NSCLC as the primary target population following the dosing of the first patient in October 2024. At the AACR Annual Meeting in April 2025, Lilly presented, on behalf of the collaboration, the clinical study design poster for the Phase 1 trial evaluating FHD-909 in patients with SMARCA4 mutated locally advanced or metastatic solid tumors who have exhausted standard treatment options. The primary target population is NSCLC. Presented synergistic preclinical data of FHD-909 in combination with pembrolizumab and KRAS inhibitors. Also at the AACR Annual Meeting in April 2025, Lilly, on behalf of the collaboration, made an oral presentation of new preclinical data demonstrating enhanced anti-tumor activity of FHD-909 in combination with standard-of-care chemotherapies, anti-PD-1 pembrolizumab and several novel KRAS inhibitors in NSCLC animal models. The combination data will inform further development plans of FHD-909. Ongoing strategic collaboration with Lilly. Collaborating with Lilly to create novel oncology medicines that includes a U.S. 50/50 co-development and co-commercialization agreement for Foghorn’s selective SMARCA2 oncology program, including a selective inhibitor and a selective degrader, and an additional undisclosed oncology target. The collaboration also includes three discovery programs from Foghorn’s proprietary Gene Traffic Control® platform. Selective CBP degrader program. Selectively targets CBP in EP300-mutated cancer cells found in many types of cancer, including bladder, gastric, and endometrial cancers. CBP and EP300 are highly similar acetyltransferases that create a synthetic lethal relationship when EP300 is mutated. Attempts to selectively drug CBP have been challenging due to the high level of similarity between the two proteins, while dual inhibition of CBP/EP300 has been limited by dose-limiting toxicities. Presented preclinical combination data with Selective degrader CBP in ER+ breast cancer. In April 2025, preclinical data showing Selective CBP degraders have combinatorial benefit with approved chemotherapies and targeted agents in solid tumors beyond EP300-mutant cancers was presented as a poster at the AACR Annual Meeting Synergistic combination activity demonstrated including with paclitaxel and CDK4/6 inhibitor abemaciclib in ER+ breast cancerFindings support combination opportunities for selective CBP degraders in solid tumors beyond EP300-mutant cancers On track for IND-enabling studies, targeting IND in 2026. Selective EP300 degrader program. Selective degradation of EP300 for the treatment of hematopoietic malignancies and prostate cancer. Attempts to selectively drug EP300 have been challenging due to the high level of similarity between EP300 and CBP, while dual inhibition of CBP/EP300 has been limited by dose limiting toxicities. EP300 lineage dependencies are established in multiple myeloma and diffuse large B cell lymphoma. Presented preclinical data showing combination with diffuse large b-cell lymphoma (DLBCL) and multiple myeloma (MM) Standard-of-Care Treatment (SoC). In April 2025, preclinical data for the Selective EP300 degrader program demonstrating biological activity in hematological malignancies was presented at the AACR Annual Meeting. Anti-proliferative activity in a broad range of hematological malignancies in vitro, including DLBCL, MM, and follicular lymphomaCombination of EP300 degrader with SoC in both DLBCL and MM are highly synergistic in vitro.Selective EP300 degradation is effective in IMiD-resistant MM cell lines. Program update expected in H2 2025. Selective ARID1B degrader program. Selectively targets and degrades ARID1B in ARID1A-mutated cancers. ARID1A is the most mutated subunit in the BAF complex and amongst the most mutated proteins in cancer. These mutations lead to a dependency on ARID1B in several types of cancer, including ovarian, endometrial, colorectal and bladder. Attempts to selectively drug ARID1B have been challenging because of the high degree of similarity between ARID1A and ARID1B and the fact that ARID1B has no enzymatic activity to target. ARID1B is a major synthetic lethal target implicated in up to 5% of all solid tumors.Developed highly potent and selective binders. Preclinical data demonstrated potent and selective small molecule binders to ARID1B.Selective degradation of ARID1B achieved. Foghorn has successfully selectively degraded ARID1B and expects to provide an update on the Selective ARID1B degrader program in H2 2025. Chromatin Biology and Degrader Platform. Foghorn continues to advance its chromatin biology and degrader platform with investments in novel ligases, long-acting injectables, oral delivery, and induced proximity. First Quarter 2025 Financial Highlights Collaboration Revenue. Collaboration revenue was $6.0 million for the three months ended March 31, 2025, compared to $5.1 million for the three months ended March 31, 2024. The increase was driven by the continued advancement of programs under the Lilly Collaboration Agreement.Research and Development Expenses. Research and development expenses were $21.6 million for the three months ended March 31, 2025, compared to $25.5 million for the three months ended March 31, 2024. The decrease is attributed to a decrease in FHD-286 costs of $2.5 million, decreases in personnel-related costs, early development and other research external costs and facilities and IT-related expenses of $2.1 million, partially offset by an increase in Lilly-partnered programs of $0.7 million.General and Administrative Expenses. General and administrative expenses were $7.2 million for the three months ended March 31, 2025, compared to $7.7 million for the three months ended March 31, 2024. This decrease was primarily due to lower consulting costs.Net Loss. Net loss was $18.8 million for the three months ended March 31, 2025, compared to a net loss of $25.0 million for the three months ended March 31, 2024.Cash, Cash Equivalents, and Marketable Securities. As of March 31, 2025, the Company had $220.6 million in cash, cash equivalents, and marketable securities, providing expected cash runway into 2027. About FHD-909FHD-909 (LY4050784) is a potent, first-in-class, allosteric, and orally available small molecule that selectively inhibits the ATPase activity of SMARCA2 (BRM) over its closely related paralog SMARCA4 (BRG1), two proteins that are the catalytic engines across all forms of the BAF complex, one of the key regulators of the chromatin regulatory system. In preclinical studies, tumors with mutations in SMARCA4 rely on SMARCA2 for their survival. FHD-909 has shown significant anti-tumor activity across multiple SMARCA4 mutant lung tumor models. About Foghorn TherapeuticsFoghorn® Therapeutics is discovering and developing a novel class of medicines targeting genetically determined dependencies within the chromatin regulatory system. Through its proprietary scalable Gene Traffic Control® platform, Foghorn is systematically studying, identifying, and validating potential drug targets within the chromatin regulatory system. The Company is developing multiple product candidates in oncology. Visit our website at www.foghorntx.com for more information on the Company, and follow us on X and LinkedIn. Forward-Looking StatementsThis press release contains “forward-looking statements.” Forward-looking statements include statements regarding the Company’s ongoing Phase 1 trial of FHD-909 in SMARCA4-mutated cancers, pre-clinical product candidates, expected timing of clinical data, expected cash runway, expected timing of regulatory filings, and research efforts and other statements identified by words such as “could,” “may,” “might,” “will,” “likely,” “anticipates,” “intends,” “plans,” “seeks,” “believes,” “estimates,” “expects,” “continues,” “projects” and similar references to future periods. Forward-looking statements are based on our current expectations and assumptions regarding capital market conditions, our business, the economy and other future conditions. Because forward-looking statements relate to the future, by their nature, they are subject to inherent uncertainties, risks and changes in circumstances that are difficult to predict. As a result, actual results may differ materially from those contemplated by the forward-looking statements. Important factors that could cause actual results to differ materially from those in the forward-looking statements include regional, national or global political, economic, business, competitive, market and regulatory conditions, including risks relating to our clinical trials and other factors set forth under the heading “Risk Factors” in the Company’s Annual Report on Form 10-K for the year ended December 31, 2024, as filed with the Securities and Exchange Commission. Any forward-looking statement made in this press release speaks only as of the date on which it is made. Condensed Consolidated Balance Sheets(In thousands) March 31, 2025 December 31, 2024Cash, cash equivalents and marketable securities$220,587 $243,747 All other assets 38,104 40,235 Total assets$258,691 $283,982 Deferred revenue, total$274,112 $280,063 All other liabilities 46,231 49,447 Total liabilities$320,343 $329,510 Total stockholders’ deficit$(61,652) $(45,528)Total liabilities and stockholders’ deficit$258,691 $283,982
Condensed Consolidated Statements of Operations(In thousands, except share and per share amounts) Three Months Ended March 31, 2025 2024 Collaboration revenue$5,952 $5,050 Operating expenses: Research and development 21,626 25,534 General and administrative 7,239 7,710 Total operating expenses$28,865 $33,244 Loss from operations$(22,913) $(28,194)Total other income, net$4,079 $3,178 Net loss$(18,834) $(25,016)Net loss per share attributable to common stockholders—basic and diluted (0.30) (0.59)Weighted average common shares outstanding—basic and diluted 62,848,673 42,428,813
Contact:Karin Hellsvik, Foghorn Therapeutics Inc.khellsvik@foghorntx.com
Phase 1Executive ChangeAACRFinancial Statement
100 Deals associated with SMARCA2 inhibitors (Arvinas LLC)
Login to view more data
R&D Status
10 top R&D records. to view more data
Login
| Indication | Highest Phase | Country/Location | Organization | Date |
|---|---|---|---|---|
| Neoplasms | Preclinical | United States | 05 Jan 2024 | |
| Neoplasms | Preclinical | United States | 05 Jan 2024 |
Login to view more data
Clinical Result
Clinical Result
Indication
Phase
Evaluation
View All Results
| Study | Phase | Population | Analyzed Enrollment | Group | Results | Evaluation | Publication Date |
|---|
No Data | |||||||
Login to view more data
Translational Medicine
Boost your research with our translational medicine data.
login
or

Deal
Boost your decision using our deal data.
login
or

Core Patent
Boost your research with our Core Patent data.
login
or
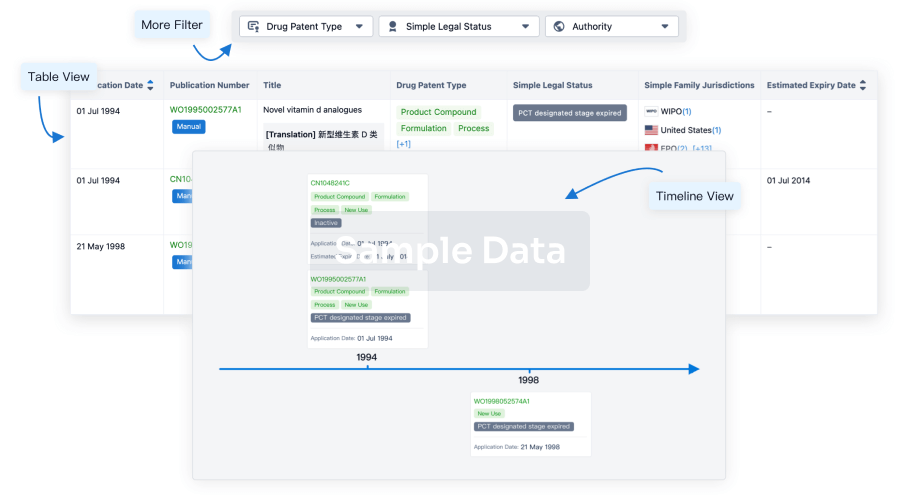
Clinical Trial
Identify the latest clinical trials across global registries.
login
or

Approval
Accelerate your research with the latest regulatory approval information.
login
or
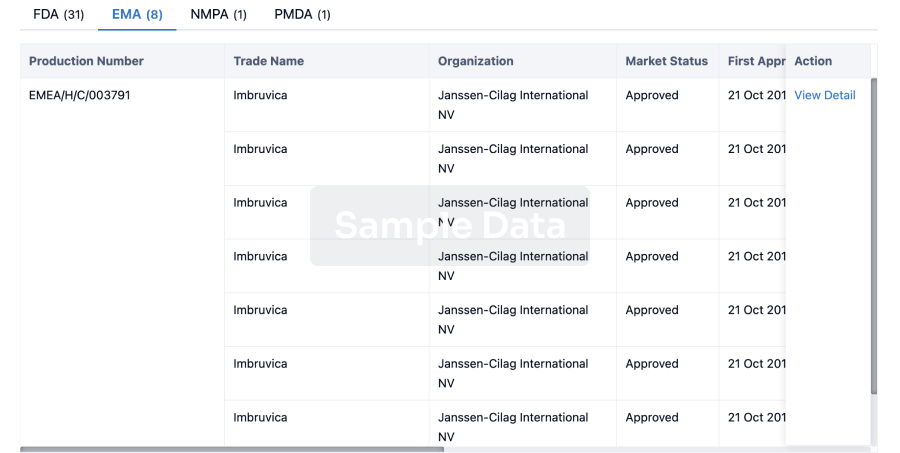
Regulation
Understand key drug designations in just a few clicks with Synapse.
login
or
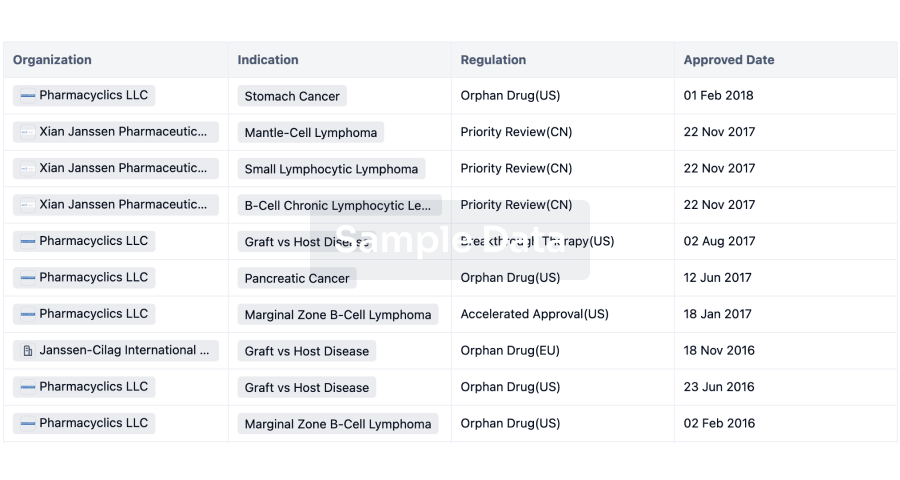
AI Agents Built for Biopharma Breakthroughs
Accelerate discovery. Empower decisions. Transform outcomes.
Get started for free today!
Accelerate Strategic R&D decision making with Synapse, PatSnap’s AI-powered Connected Innovation Intelligence Platform Built for Life Sciences Professionals.
Start your data trial now!
Synapse data is also accessible to external entities via APIs or data packages. Empower better decisions with the latest in pharmaceutical intelligence.
Bio
Bio Sequences Search & Analysis
Sign up for free
Chemical
Chemical Structures Search & Analysis
Sign up for free
