Request Demo
Last update 23 Jan 2025
ATP6V1F
Last update 23 Jan 2025
Basic Info
Synonyms ATP6S14, ATP6V1F, ATPase H+ transporting V1 subunit F + [7] |
Introduction Subunit of the V1 complex of vacuolar(H+)-ATPase (V-ATPase), a multisubunit enzyme composed of a peripheral complex (V1) that hydrolyzes ATP and a membrane integral complex (V0) that translocates protons (PubMed:33065002). V-ATPase is responsible for acidifying and maintaining the pH of intracellular compartments and in some cell types, is targeted to the plasma membrane, where it is responsible for acidifying the extracellular environment (By similarity). |
Analysis
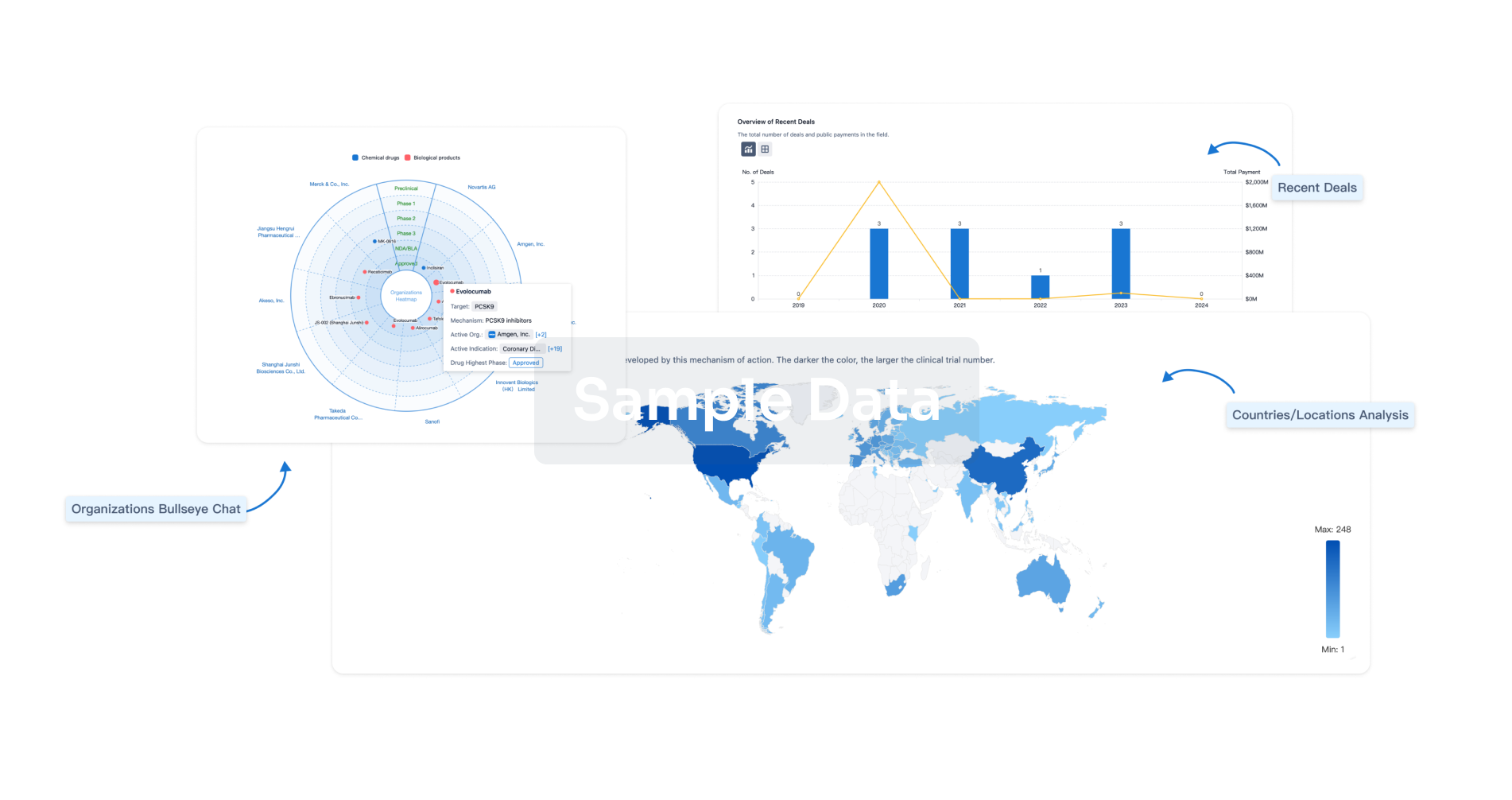
Perform a panoramic analysis of this field.
login
or
Chat with Hiro
Get started for free today!
Accelerate Strategic R&D decision making with Synapse, PatSnap’s AI-powered Connected Innovation Intelligence Platform Built for Life Sciences Professionals.
Start your data trial now!
Synapse data is also accessible to external entities via APIs or data packages. Empower better decisions with the latest in pharmaceutical intelligence.
Bio
Bio Sequences Search & Analysis
Sign up for free
Chemical
Chemical Structures Search & Analysis
Sign up for free