Last update 01 Nov 2024
GINS2
Last update 01 Nov 2024
Basic Info
Synonyms DNA replication complex GINS protein PSF2, GINS complex subunit 2, GINS2 + [2] |
Introduction Required for correct functioning of the GINS complex, a complex that plays an essential role in the initiation of DNA replication, and progression of DNA replication forks (PubMed:17417653). GINS complex is a core component of CDC45-MCM-GINS (CMG) helicase, the molecular machine that unwinds template DNA during replication, and around which the replisome is built (PubMed:32453425, PubMed:34694004, PubMed:34700328, PubMed:35585232). |
Analysis
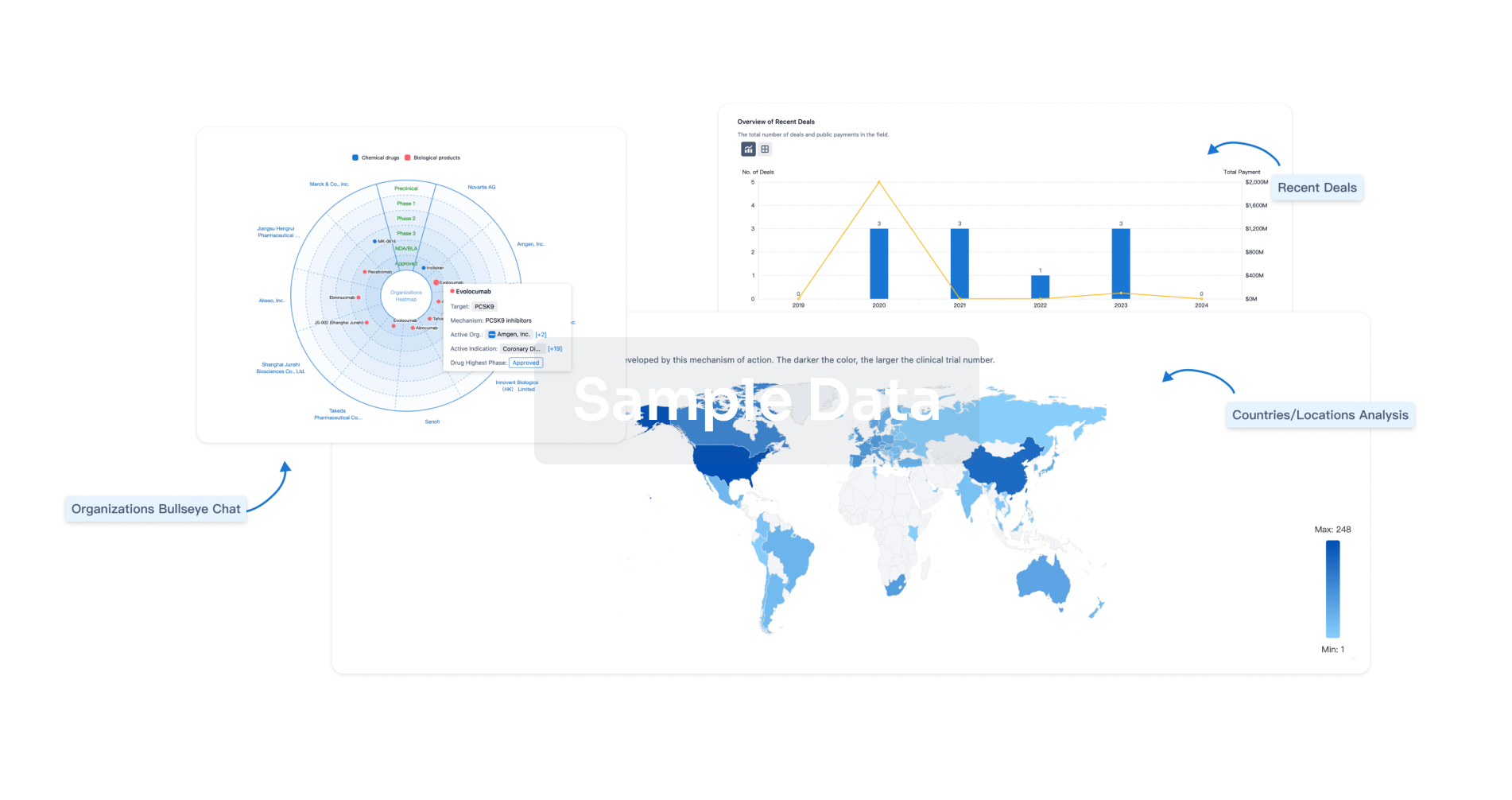
Perform a panoramic analysis of this field.
login
or
Chat with Hiro
Get started for free today!
Accelerate Strategic R&D decision making with Synapse, PatSnap’s AI-powered Connected Innovation Intelligence Platform Built for Life Sciences Professionals.
Start your data trial now!
Synapse data is also accessible to external entities via APIs or data packages. Empower better decisions with the latest in pharmaceutical intelligence.
Bio
Bio Sequences Search & Analysis
Sign up for free
Chemical
Chemical Structures Search & Analysis
Sign up for free
