Request Demo
Last update 08 May 2025
ACTG2
Last update 08 May 2025
Basic Info
Synonyms ACTA3, ACTG2, actin gamma 2, smooth muscle + [7] |
Introduction Actins are highly conserved proteins that are involved in various types of cell motility and are ubiquitously expressed in all eukaryotic cells. |
Related
1
Drugs associated with ACTG2Target |
Mechanism ACTG2 inhibitors |
Active Org. |
Originator Org. |
Active Indication |
Inactive Indication- |
Drug Highest PhasePreclinical |
First Approval Ctry. / Loc.- |
First Approval Date20 Jan 1800 |
100 Clinical Results associated with ACTG2
Login to view more data
100 Translational Medicine associated with ACTG2
Login to view more data
0 Patents (Medical) associated with ACTG2
Login to view more data
195
Literatures (Medical) associated with ACTG201 Jun 2025·Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease
Overexpression of LMOD1 induces oxidative stress and enhances cell apoptosis of melanoma through the RIG-I like receptor pathway
Article
Author: Lei, Hua ; Wan, Huiying ; Huang, Linxue ; Chen, Mingyi
01 Apr 2025·Journal of Biological Chemistry
Declining activity of serum response factor in aging aorta in relation to aneurysm progression
Article
Author: Bastrup, Joakim Armstrong ; Swärd, Karl ; Holmberg, Johan ; Arévalo Martinez, Marycarmen ; Kawka, Katarzyna ; Rippe, Catarina ; Jepps, Thomas A ; Albinsson, Sebastian
01 Jan 2025·Computational and Structural Biotechnology Journal
Dynamical features of smooth muscle actin pathological mutants: The arginine-257(258)-Cysteine cases
Article
Author: de Rosa, M ; Cavalli, A ; Chiappori, F ; Viti, F ; Palma, F Di
Analysis
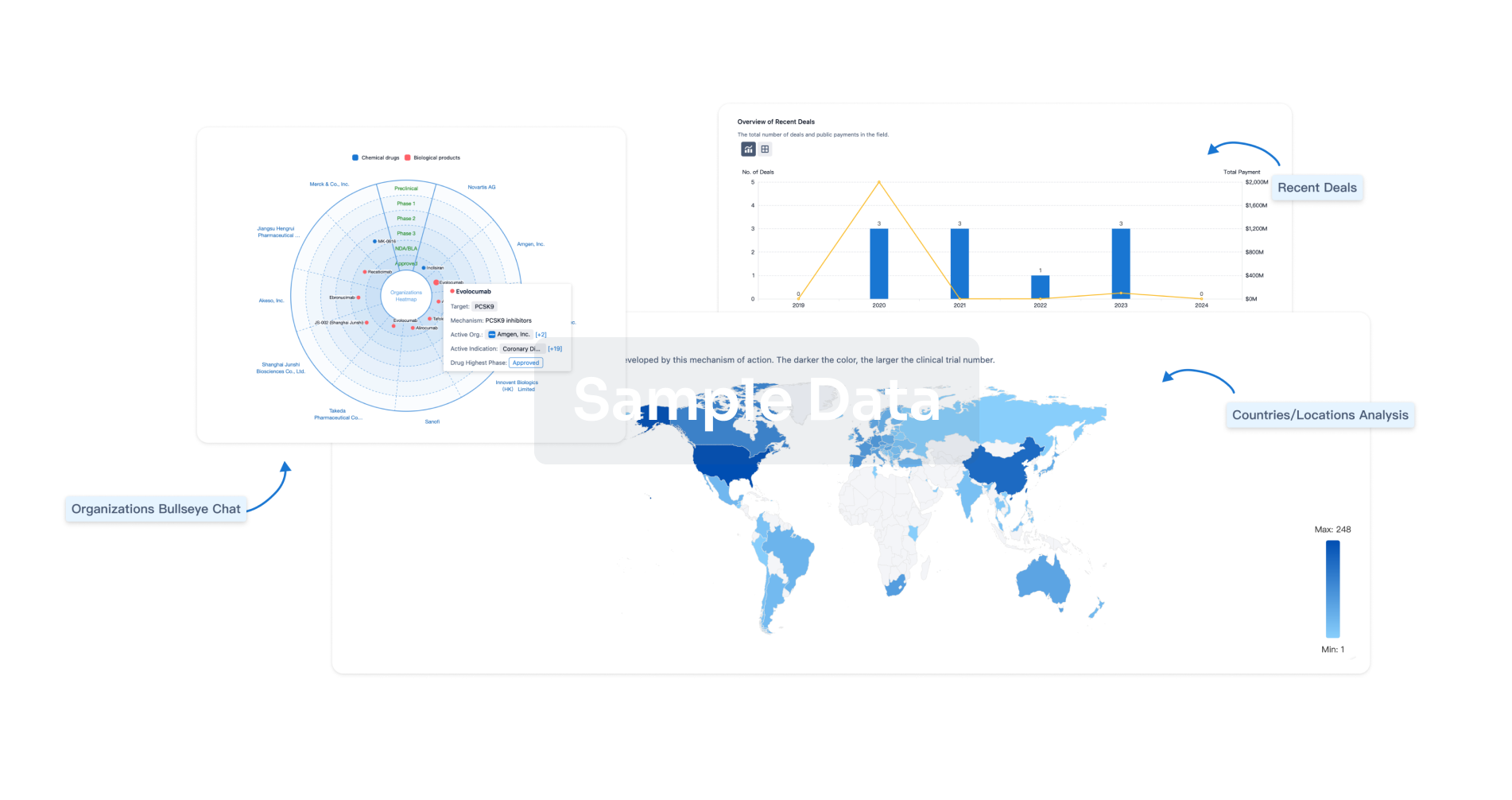
Perform a panoramic analysis of this field.
login
or
AI Agents Built for Biopharma Breakthroughs
Accelerate discovery. Empower decisions. Transform outcomes.
Get started for free today!
Accelerate Strategic R&D decision making with Synapse, PatSnap’s AI-powered Connected Innovation Intelligence Platform Built for Life Sciences Professionals.
Start your data trial now!
Synapse data is also accessible to external entities via APIs or data packages. Empower better decisions with the latest in pharmaceutical intelligence.
Bio
Bio Sequences Search & Analysis
Sign up for free
Chemical
Chemical Structures Search & Analysis
Sign up for free
