Request Demo
Last update 08 May 2025
HPX
Last update 08 May 2025
Basic Info
Synonyms Beta-1B-glycoprotein, hemopexin, HPX |
Introduction Binds heme and transports it to the liver for breakdown and iron recovery, after which the free hemopexin returns to the circulation. |
Related
1
Drugs associated with HPXTarget |
Mechanism HPX modulators |
Active Org. |
Originator Org. |
Active Indication |
Inactive Indication- |
Drug Highest PhasePhase 2/3 |
First Approval Ctry. / Loc.- |
First Approval Date20 Jan 1800 |
2
Clinical Trials associated with HPXNCT06699849
A Phase 2/Phase 3, Multicenter, Randomized, Multiple-Dose, Double-Blind, Placebo-Controlled Adaptive Study to Evaluate the Safety, Efficacy, and Pharmacokinetics of CSL889 in Adults and Adolescents With Sickle Cell Disease During Vaso-Occlusive Crisis
This study consists of two parts: phase 2 (Part A) and phase 3 (Part B). It is a multicenter study designed to evaluate the safety, effectiveness, and pharmacokinetics (PK) of CSL889 (human hemopexin) when given intravenously (IV) to adults and adolescents with sickle cell disease (SCD) experiencing vaso-occlusive crises (VOC). The main objectives of the study are to assess how CSL889 affects the time it takes for VOC to resolve in participants with SCD, and to evaluate the safety and tolerability of CSL889 in study participants.
Start Date14 Feb 2025 |
Sponsor / Collaborator |
NCT04285827
A 2-Part, Phase 1, Multi-Center, Single-Dose, Open Label, Study to Evaluate the Safety, Tolerability, and Pharmacokinetics of CSL889 in Adult Patients With Sickle Cell Disease
This is a phase 1, first-in-human, multi-center, open-label, single dose cohort study to evaluate the safety and tolerability, pharmacokinetics (PK), exploratory pharmacodynamics (PD), and biomarkers of target engagement of CSL889 following single intravenous (IV) doses in subjects with sickle cell disease (SCD). The study involves sequential dose escalation of cohorts with between-group assessments of key safety and PK variables.
Start Date20 May 2021 |
Sponsor / Collaborator |
100 Clinical Results associated with HPX
Login to view more data
100 Translational Medicine associated with HPX
Login to view more data
0 Patents (Medical) associated with HPX
Login to view more data
1,343
Literatures (Medical) associated with HPX31 Dec 2025·Hematology
Potential role of hemopexin in venous thromboembolism (VTE) mediated through effects on apoptosis-related proteins
Article
Author: Yu, Xiao-hui ; Tang, Peng ; Liu, Hua-gang ; Zhuang, Jun-li ; Zhou, Jiang-qiao ; Li, Jie ; Deng, Hong-ping ; Zhang, Zhong-jun
01 Jun 2025·Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease
Multiple classes of human intracellular Heme-binding proteins with pathology-associated polymorphisms of heme coordinating residues
Review
Author: Tsiftsoglou, Stefanos A ; Tsiftsoglou, Asterios S
01 Jun 2025·Life Sciences
Decoding Proteomic cross-talk between hypobaric and normobaric hypoxia: Integrative analysis of oxidative stress, cytoskeleton remodeling, and inflammatory pathways
Article
Author: Mohanty, Swaraj ; Ahmad, Yasmin ; Sharma, Poornima
Analysis
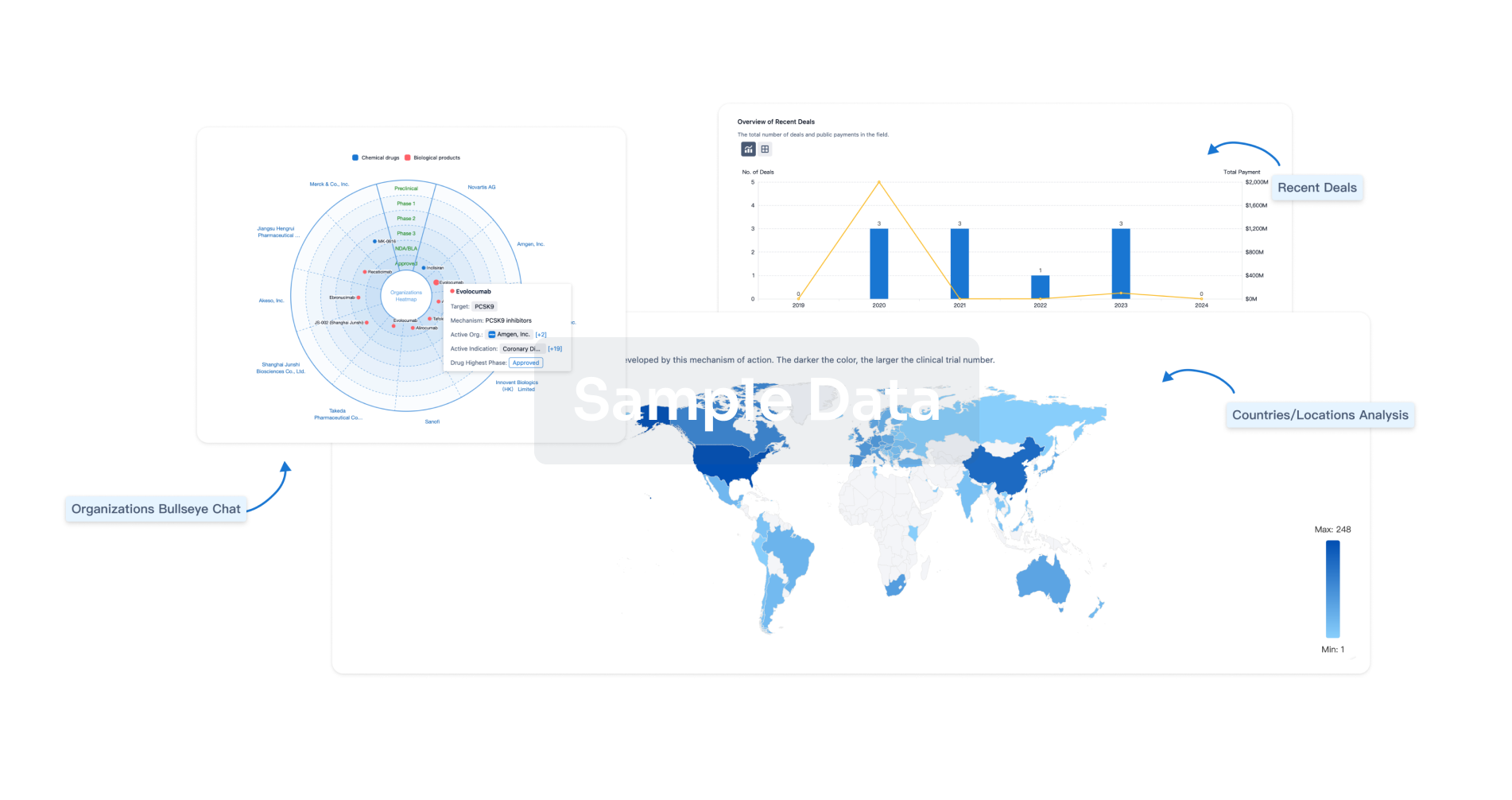
Perform a panoramic analysis of this field.
login
or
AI Agents Built for Biopharma Breakthroughs
Accelerate discovery. Empower decisions. Transform outcomes.
Get started for free today!
Accelerate Strategic R&D decision making with Synapse, PatSnap’s AI-powered Connected Innovation Intelligence Platform Built for Life Sciences Professionals.
Start your data trial now!
Synapse data is also accessible to external entities via APIs or data packages. Empower better decisions with the latest in pharmaceutical intelligence.
Bio
Bio Sequences Search & Analysis
Sign up for free
Chemical
Chemical Structures Search & Analysis
Sign up for free
