Durvalumab: Sequence Analysis and Clinical Application Prospects of Immune Checkpoint Inhibitors
Checkpoint inhibitors have emerged as a major focus in cancer therapy, demonstrating remarkable efficacy by activating or enhancing the immune system's ability to fight tumors. These inhibitors, primarily targeting checkpoints such as PD-1/PD-L1 and CTLA-4, work by reversing tumor-induced immunosuppression, activating T cells, and improving their capacity to identify and eliminate cancer cells. Among the many immune checkpoint inhibitors, Durvalumab (brand name Imfinzi) stands out as particularly noteworthy.
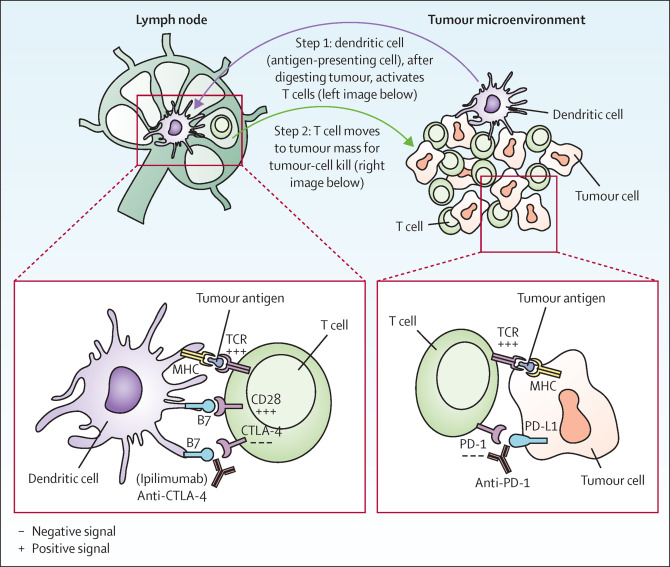
Durvalumab’s anti-cancer mechanism involves blocking the interaction between PD-L1 on tumor cells and the PD-1 receptor on T cells. This disruption restores T-cell recognition and attack capabilities against tumors. Additionally, Durvalumab boosts adaptive immune responses and is often used in combination with other therapies, such as radiotherapy or chemotherapy, to enhance treatment efficacy.
Approved by the U.S. Food and Drug Administration (FDA) in May 2017, Durvalumab became the second PD-L1 inhibitor to receive approval. Initially indicated for locally advanced or metastatic urothelial carcinoma, it has since gained approval in multiple countries and regions for treating several cancer types, including non-small cell lung cancer (NSCLC), extensive-stage small cell lung cancer (ES-SCLC), bile duct cancer, and unresectable hepatocellular carcinoma.
In the development and application of antibody-based therapies like Durvalumab, amino acid sequences play a critical role. They determine the three-dimensional structure, specificity, and biological functionality of antibodies. Database queries, comparisons, and analyses of monoclonal antibody amino acid sequences are crucial for understanding their mechanisms of action, optimizing drug design, predicting therapeutic effects, and improving efficacy and safety.
Furthermore, databases also aid in monitoring patent trends, avoiding intellectual property conflicts, accelerating drug development processes, and fostering innovation in the pharmaceutical industry.
Patsnap Bio offers researchers an invaluable platform for accessing detailed antibody amino acid sequences and related drug patent data. This enables in-depth analysis of molecular structure and functionality while supporting drug design, efficacy evaluation, and the formulation of clinical strategies. Patsnap Bio also helps mitigate intellectual property risks, ensuring compliance during the R&D process, and facilitates smoother market entry and commercialization.
Using Patsnap Bio, researchers can quickly retrieve Durvalumab’s sequence data. This step-by-step process highlights the database's utility in analyzing antibody drugs, paving the way for improved drug design, safety, and market readiness.
Step 1: Retrieve the Amino Acid Sequence of the Drug
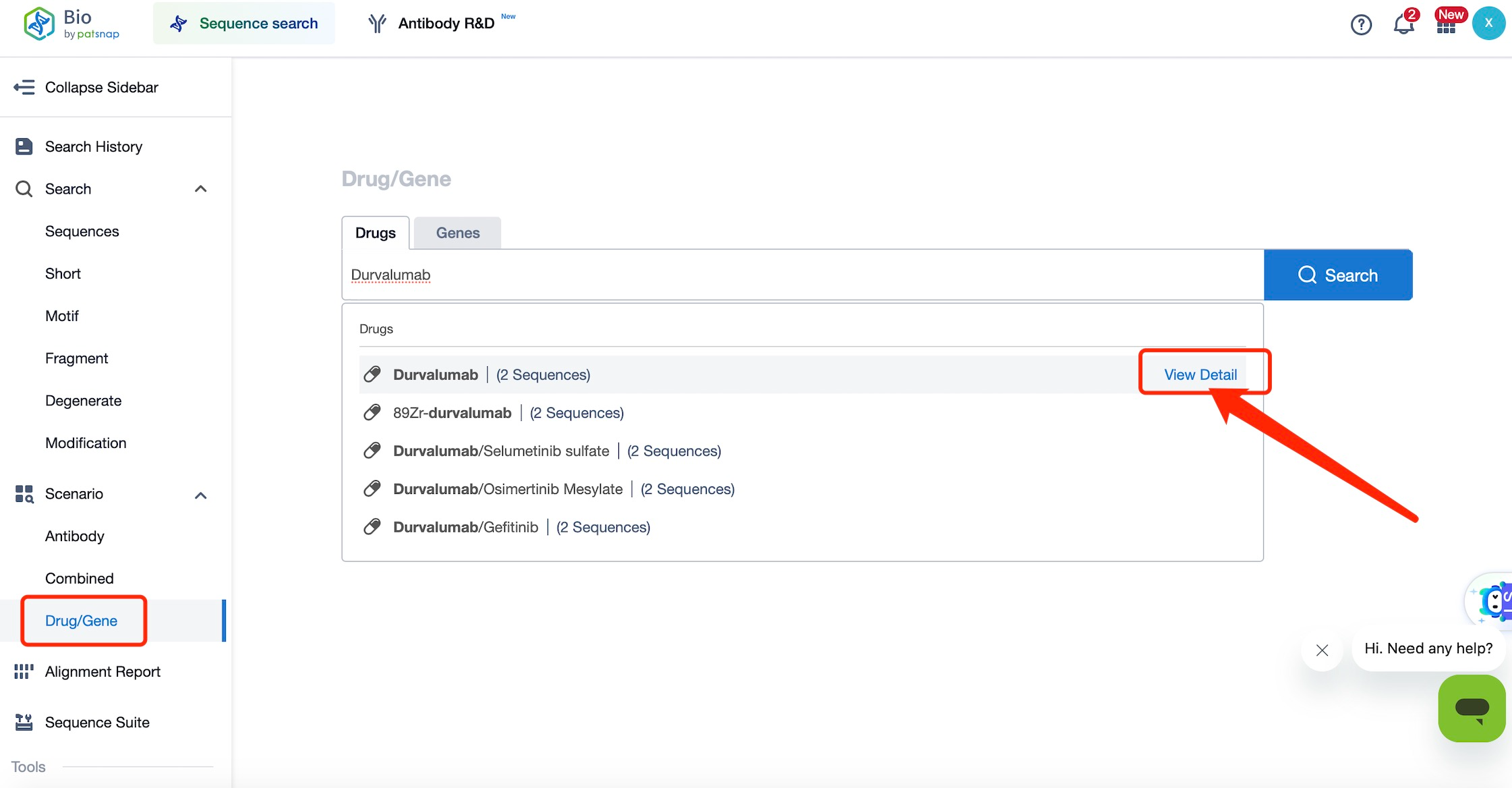
First, log in to Patsnap Bio. On the homepage, if you do not know the exact sequence information of Durvalumab, you can input the drug name "Durvalumab" in the Drug/Gene Index on the left sidebar and click search.
Step 2: Analyze the Sequence
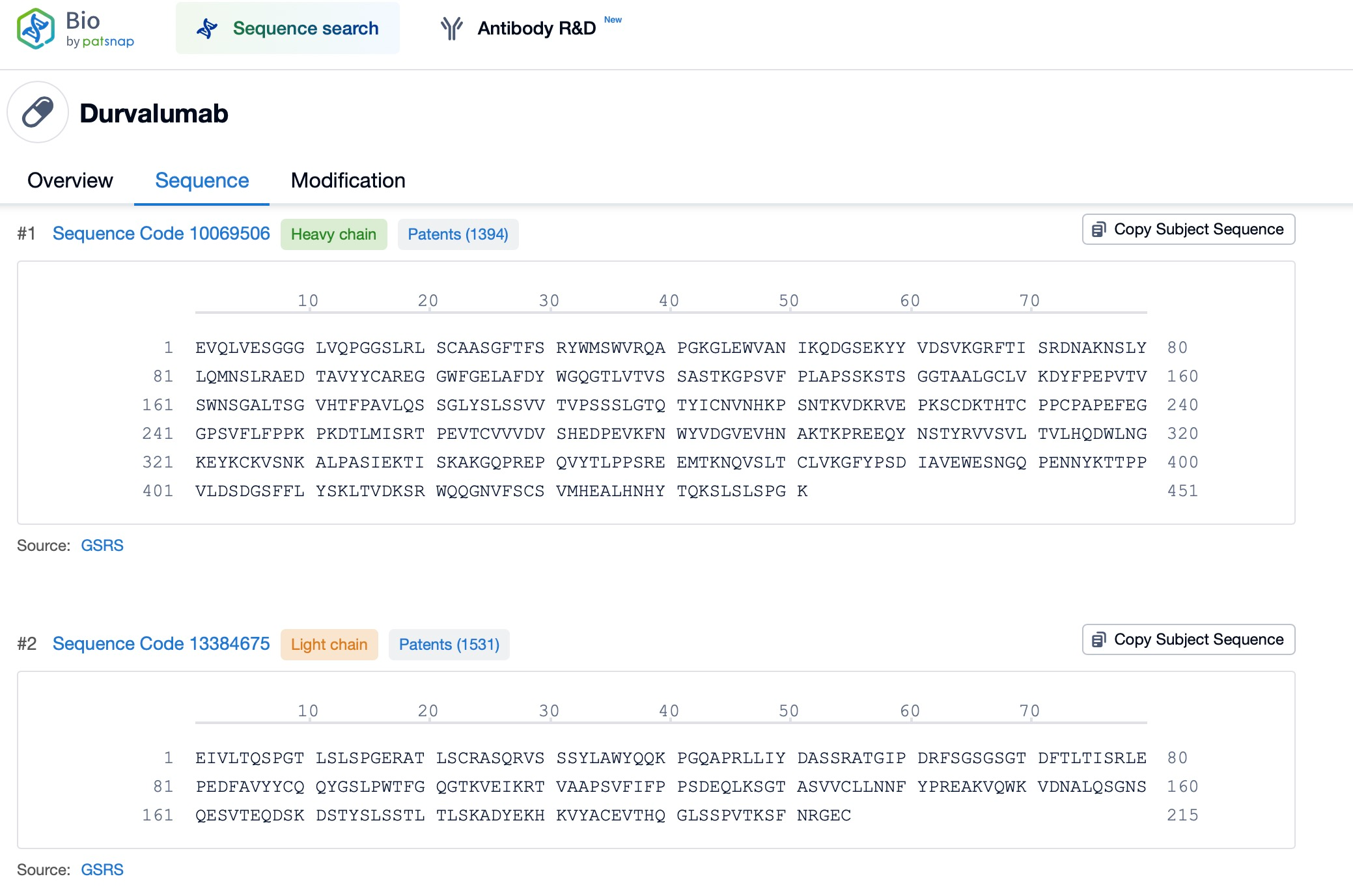
Through the search, two sequence data entries for Durvalumab were identified: the antibody light chain, which consists of 215 amino acids, and the antibody heavy chain, which consists of 451 amino acids.
Durvalumab contains two functional domains: a variable domain (VL) and a constant domain (CL). The variable domain of the light chain is responsible for specific antigen binding, while the constant domain is involved in the antibody's effector functions, such as activating the complement system or binding to surface receptors on immune cells. The variable region (VL) of the light chain exhibits high variability, enabling it to bind to a wide range of antigen epitopes. In contrast, the constant region (CL) is relatively conserved across antibodies of the same isotype, and while it does not directly participate in antigen binding, it plays an essential role in antibody classification and functionality.
The variable region (VL) of Durvalumab's light chain contains three complementarity-determining regions (CDRs), which are the critical regions of the antibody molecule that interact with antigens. The diversity in the CDR sequences enables antibodies to recognize and bind to a wide variety of antigen epitopes. Within the variable region of the antibody light chain, CDR1, CDR2, and CDR3 collectively form the antigen-binding site, which interacts with antigen epitopes to achieve specific recognition. The amino acid sequence and structure of the CDRs are highly diverse across different antibodies, making them one of the primary sources of antibody diversity. The precise definition and analysis of CDR regions are crucial to understanding the interactions between antibodies and antigens.
Various researchers have proposed multiple CDR definition schemes, such as Kabat, Chothia, and IMGT, which define the boundaries of CDRs based on sequence or structural comparisons. Notably, the International ImMunoGeneTics Information System (IMGT) has introduced a standardized numbering system applicable to all protein sequences within the immunoglobulin superfamily. The IMGT scheme defines CDRs and framework regions comprehensively based on alignments of germline V-gene amino acid sequences.
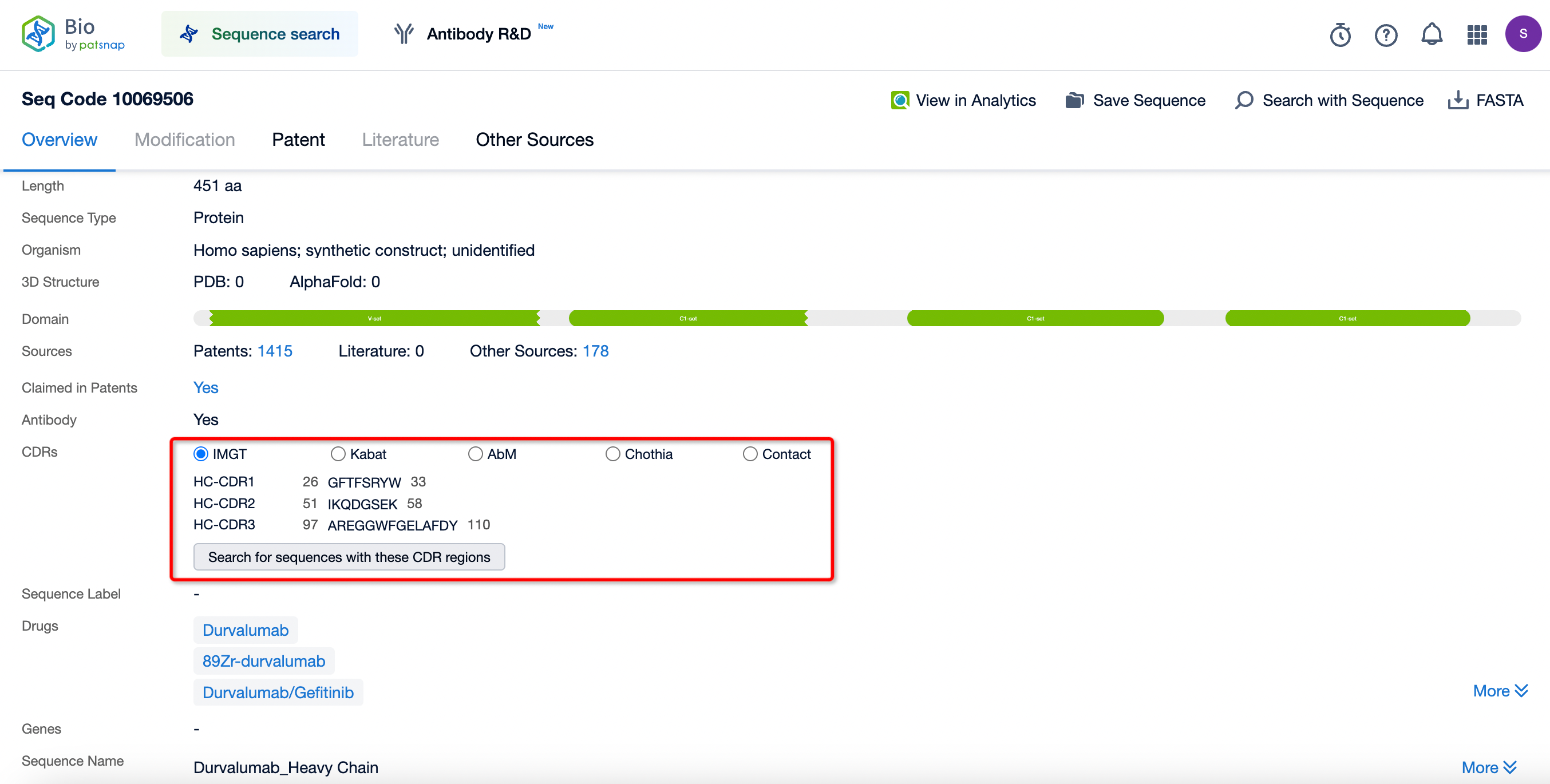
The heavy chain of Durvalumab consists of one variable domain (V-set) and three constant domains (C1-set), among which the constant domains are responsible for the antibody's effector functions. These include binding to Fc receptors on immune cells, thereby triggering antibody-dependent cell-mediated cytotoxicity (ADCC), promoting phagocytosis, and activating the complement system. These mechanisms significantly enhance the body's immune response against tumor cells.
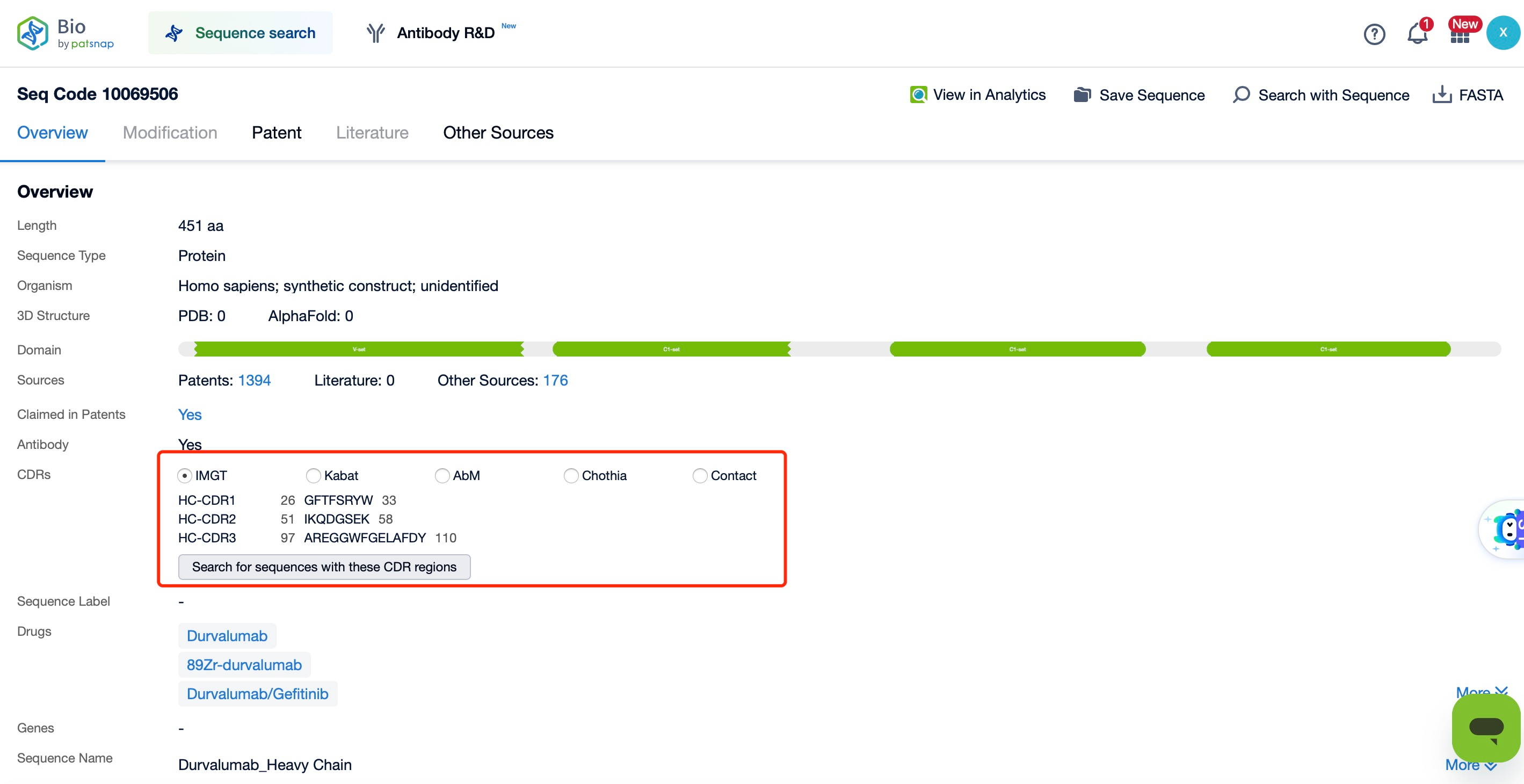
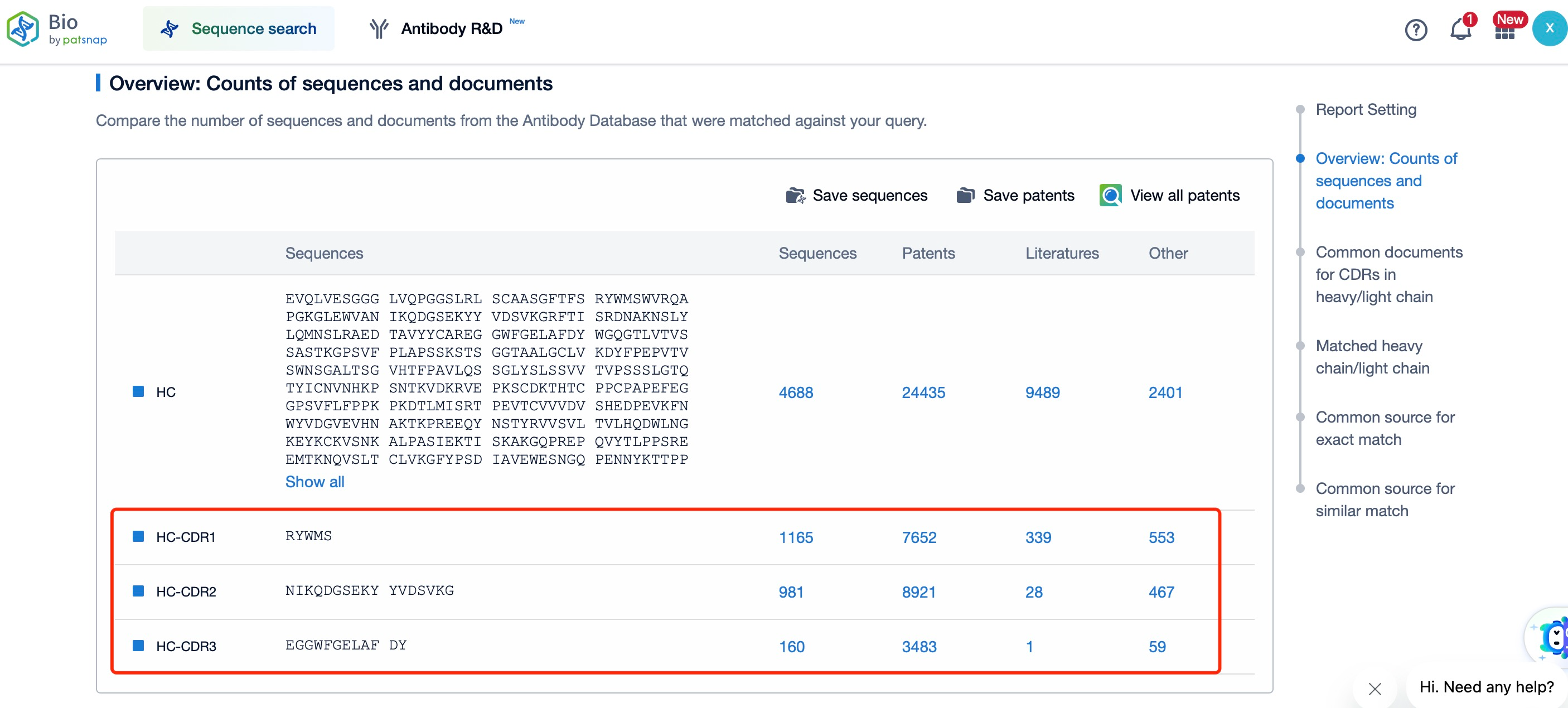
Through Patsnap Bio, we found that although the CDR regions of the heavy and light chains of Durvalumab are structurally similar, their sequences are entirely different. This is because variations in the CDRs are critical to antibody diversity, which forms the basis of the adaptive immune response. Each CDR region has a unique sequence, allowing different antibody molecules to recognize different antigens. This high variability is generated during B cell development through somatic hypermutation and antibody gene rearrangement.
Moreover, using Patsnap Bio, we can easily retrieve the sequences of the heavy and light chains of Durvalumab, as well as their CDR sequences. This allows users to identify competing products within the same therapeutic area, analyze patent landscapes focused on the same target antibodies, assess clinical outcomes, explore research and development trends, and screen researchers and institutions working on similar projects. This facilitates close monitoring of the latest industry data insights and enables professionals to make more informed decisions in biopharmaceutical R&D and intellectual property (IP) management.
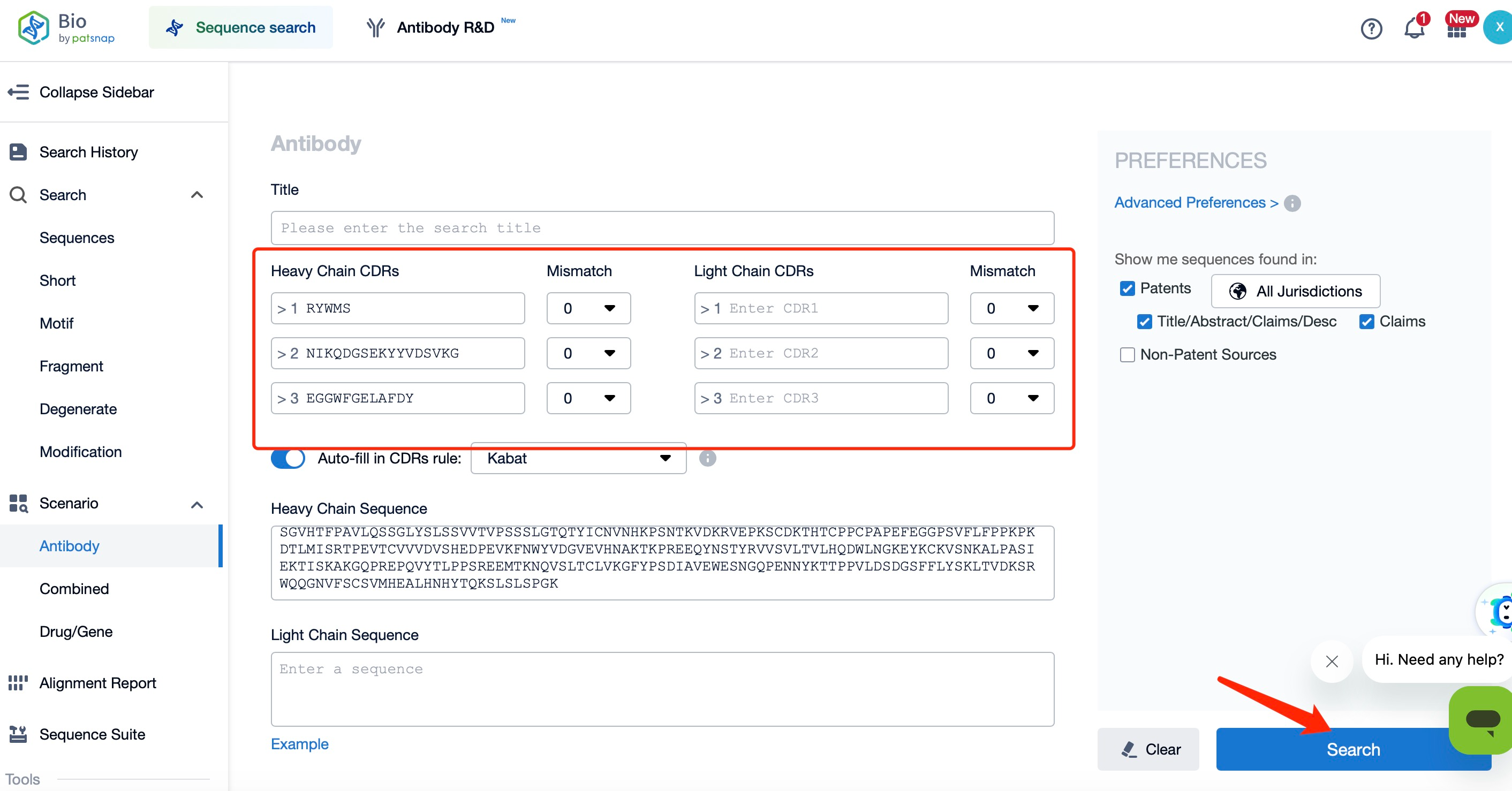
Patsnap Bio also offers a Generalized Sequence Search function, which is the world's first generalized sequence alignment algorithm. It supports the retrieval of "trillions of sequences" hidden in patents, helping to avoid the risk of sequence omission in freedom-to-operate (FTO) analysis. This feature is particularly valuable for evaluating the patentability of sequence-based innovations and performing FTO assessments, ensuring that research outcomes do not infringe on others' patent rights while also preventing others from infringing on your own intellectual property.
Summary
As an immune checkpoint inhibitor, Durvalumab has demonstrated significant efficacy in the treatment of various cancers, including non-small cell lung cancer (NSCLC), small cell lung cancer (SCLC), urothelial carcinoma, and biliary tract cancer. Its therapeutic potential extends beyond monotherapy and includes combination strategies with other anticancer agents, such as olaparib, bevacizumab, and platinum-based chemotherapy. The amino acid sequence of Durvalumab, particularly its variable regions and complementary-determining regions (CDRs), is key to its specificity and efficacy. Through in-depth analyses using databases like Patsnap Bio, users can comprehensively retrieve and compare antibody drug sequences, including essential CDR information. These data are critical for elucidating the mechanisms of action of antibody therapeutics, optimizing drug design, predicting efficacy, and improving safety profiles. Additionally, patent information provided by the database supports the monitoring of competitive landscapes within the field of the same target antibodies, and helps assess IP risks for new drug development. This enables strategic decision-making during the early stages of drug development. Patsnap Bio’s Generalized Sequence Search function further enhances its application in biopharmaceutical R&D and IP management, offering a crucial tool for thoroughly evaluating the patentability of R&D outcomes.
Better answers for better bio-innovations!
Validate novelty, eliminate risk, and innovate with confidence using the world’s largest sequence database curated from millions of patent and non-patent sources.
Patsnap Bio helps you turn weeks into minutes with cutting-edge AI-enabled tools built to master the complexities of sequence retrieval and automate IP analysis with precision and ease.
With best-in-class coverage of protein and nucleic acid sequences combined with state-of- the-art search algorithms, you’ll spend less time searching and more time bringing your bio-innovations to market.
Reference
Dall'Olio, F. G. et al. Tumour burden and efficacy of immune-checkpoint inhibitors. Nat Rev Clin Oncol 19, 75-90 (2022). https://doi.org:10.1038/s41571-021-00564-3