Application of Machine Learning in ADC Drug Linker Design
In the development of antibody-drug conjugates (ADCs), selecting an appropriate linker is a critical factor that determines the efficacy and safety of the final product. Traditional methods of linker screening often rely on time-consuming and expensive laboratory tests, whereas modern machine learning (ML) techniques provide more efficient alternatives. By collecting large datasets from literature, patents, and public databases and using advanced data preprocessing and feature selection techniques, researchers can identify features closely related to linker stability. These features include molecular structure, functional groups, molecular weight, solubility, and pKa values, which form the basis for subsequent machine learning model training.
AI-Based ADC Linker Simulation and Prediction
In ADC development, choosing the right linker is crucial to the product’s efficacy and safety. Machine learning algorithms simulate and predict the interactions between different types of linkers, antibodies, and cytotoxic drugs, but this requires extensive data preparation. First, a large amount of data on linkers, antibodies, and cytotoxic drugs is collected from existing literature, patents, and public databases, followed by preprocessing to extract features related to linker stability, such as molecular structure, functional groups, molecular weight, solubility, and pKa. Then, feature selection techniques (e.g., PCA, LASSO) filter out the most significant features that impact linker stability, and machine learning algorithms (e.g., Random Forests, Support Vector Machines, Neural Networks) are used to train models to predict linker stability under various conditions.
With a well-trained model, simulations of interactions between linkers, antibodies, and cytotoxic drugs can be conducted, and the stability of these combinations can be evaluated under different environmental conditions (e.g., pH changes, temperature variations, oxidative-reductive conditions). Additionally, molecular dynamics simulation software can be used to simulate structural changes in linkers under different conditions, further evaluating their stability. Quantum chemical methods (e.g., Density Functional Theory, DFT) can also be used to calculate bond energies and charge distributions, predicting the stability of linkers in chemical reactions.
AI-Driven Efficient Screening of ADC Drug Linkers
Efficient screening leverages deep learning and other advanced technologies to accelerate the identification of candidate linkers with ideal chemical and biological stability. While traditional methods rely on labor-intensive and costly experiments, modern computational techniques provide more effective alternatives. Through deep learning models, researchers can analyze structural and functional features of linker molecules from large datasets, allowing for the prediction of linkers that are more likely to meet the required stability standards.
With the aid of deep learning, a large number of potential linker molecules can be rapidly evaluated on computers, significantly reducing screening time. These models can be trained on known linkers and their related properties to automatically identify molecules that are stable chemically and biologically. Furthermore, the models can optimize candidate materials by simulating their behavior under various physiological conditions, ensuring that they remain stable in real-world applications.
The efficient screening process is not limited to static predictions but can also explore the dynamic behavior of linkers under physiological conditions using molecular dynamics simulations and quantum chemical calculations. Deep learning models excel at learning the structure-function relationships of linkers from complex datasets, enabling researchers to quickly assess potential linker candidates and optimize their chemical and biological stability.
In personalized medicine, AI technology’s application goes beyond improving diagnostic accuracy and extends to the drug design stage. By analyzing patients’ genomic, proteomic, and other omics data, AI can identify biomarkers associated with drug response and optimize linker design accordingly. For example, in cancer therapy, AI can predict a patient’s response to ADCs with different linkers based on genetic characteristics, thereby recommending the most effective treatment. This highly personalized treatment strategy not only increases the specificity and efficacy of the therapy but also reduces unnecessary side effects.
AI Aids in Optimizing ADC Drug Linker Design
In terms of design optimization, AI and big data analysis can significantly enhance the efficiency and effectiveness of linker design for ADCs. By analyzing vast amounts of structural data and biological activity information, AI can identify linker properties that enable ADCs to remain stable in the bloodstream while ensuring the effective release of cytotoxic drugs within the tumor microenvironment.
AI can assist in designing linkers with the appropriate length and chemical composition to ensure their stability in the blood. Through specific mechanisms such as pH sensitivity, enzyme sensitivity, or reduction sensitivity, these linkers can be programmed to break down at the right moment inside or near tumor cells, releasing the cytotoxic payload. This ensures the drug's integrity during transport while achieving precise release at the target site, minimizing damage to healthy tissues. Additionally, AI can help design chemical properties of the linker, such as the balance between hydrophilicity and hydrophobicity, to improve the pharmacokinetic profile of the ADC, ensuring optimal drug distribution and elimination in the body. In this way, AI not only enhances the precision of linker design but also boosts the overall therapeutic efficacy and safety of ADCs.
AI Assists in the Field of Personalized Medicine
In personalized medicine, AI's applications go far beyond improving diagnostic accuracy. By analyzing patient-specific information such as genomic data, lifestyle, and medical history, AI can provide tailored treatment recommendations for each individual. Especially in customized drug development, AI can help identify linker structures suitable for specific patient groups, ensuring the best therapeutic outcomes at the individual level.
With advanced algorithms like deep learning, AI can process and analyze large volumes of medical data, including but not limited to multi-omics data such as genomics, proteomics, and metabolomics, as well as clinical information like electronic health records and imaging data. Based on this data, AI can identify biomarkers related to drug response and subsequently optimize linker design. This allows the cytotoxic drug to be effectively released at the tumor site while maintaining stability in the bloodstream, thus minimizing harm to healthy tissues. For instance, in cancer therapy, AI can analyze the genetic profile of a patient’s tumor and predict their response to different ADCs with varying linkers, thereby recommending the most effective treatment plan. This approach not only increases the precision and efficacy of the treatment but also reduces unnecessary side effects, improving patients' quality of life and treatment success rates.
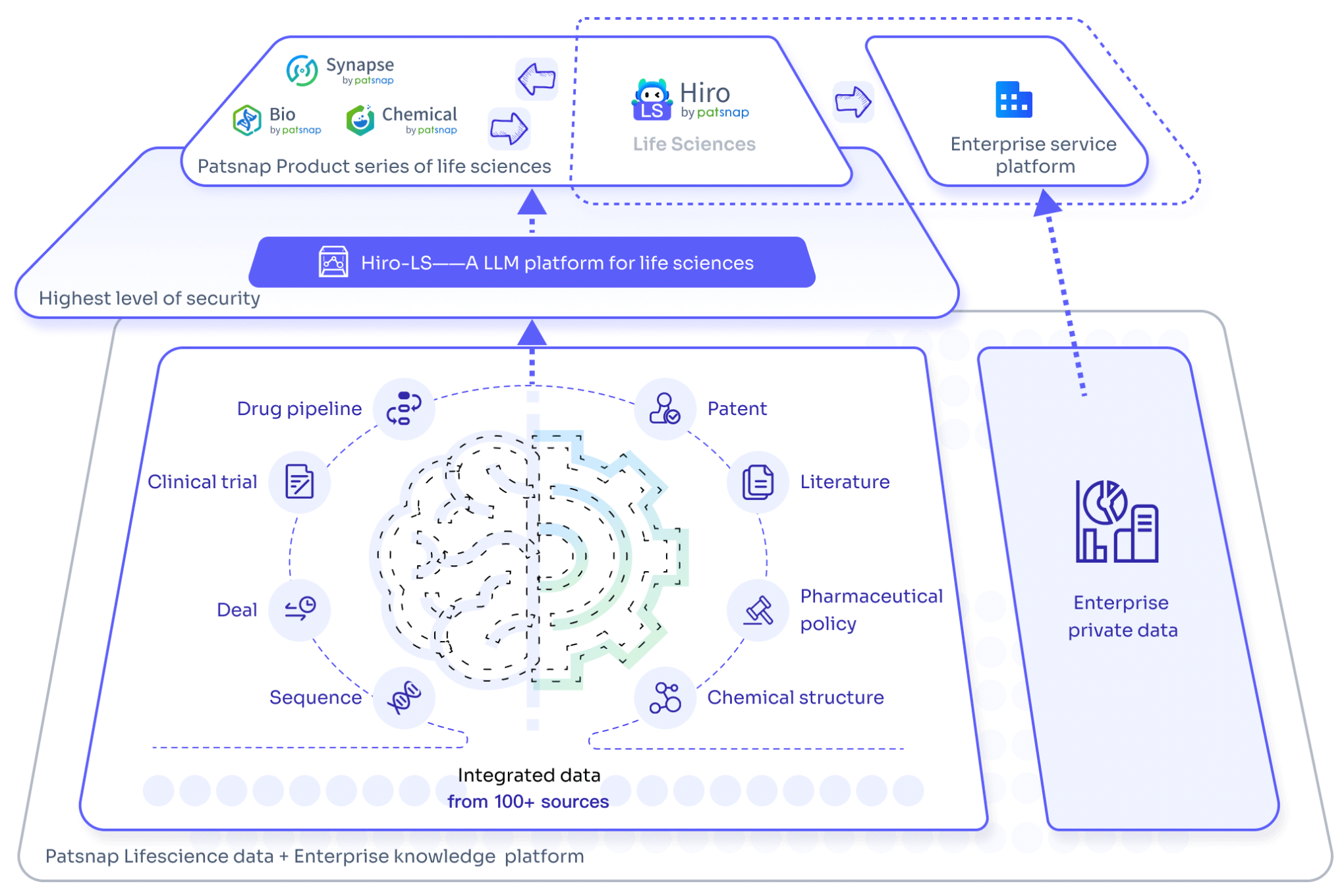
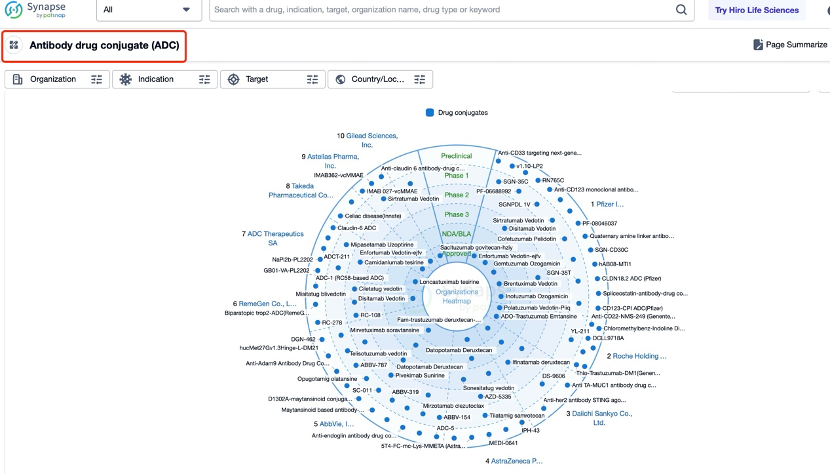
ADCs Screening with Patsnap Synapse
Patsnap Synapse plays a vital role in the development of ADCs by integrating a comprehensive range of global drug data, patent information, clinical trial results, and literature resources. This powerful platform provides researchers with tools to effectively search and filter through drug-related information, including the ability to refine results based on target, drug type, and other relevant attributes.
Antibody Screening with Patsnap Bio
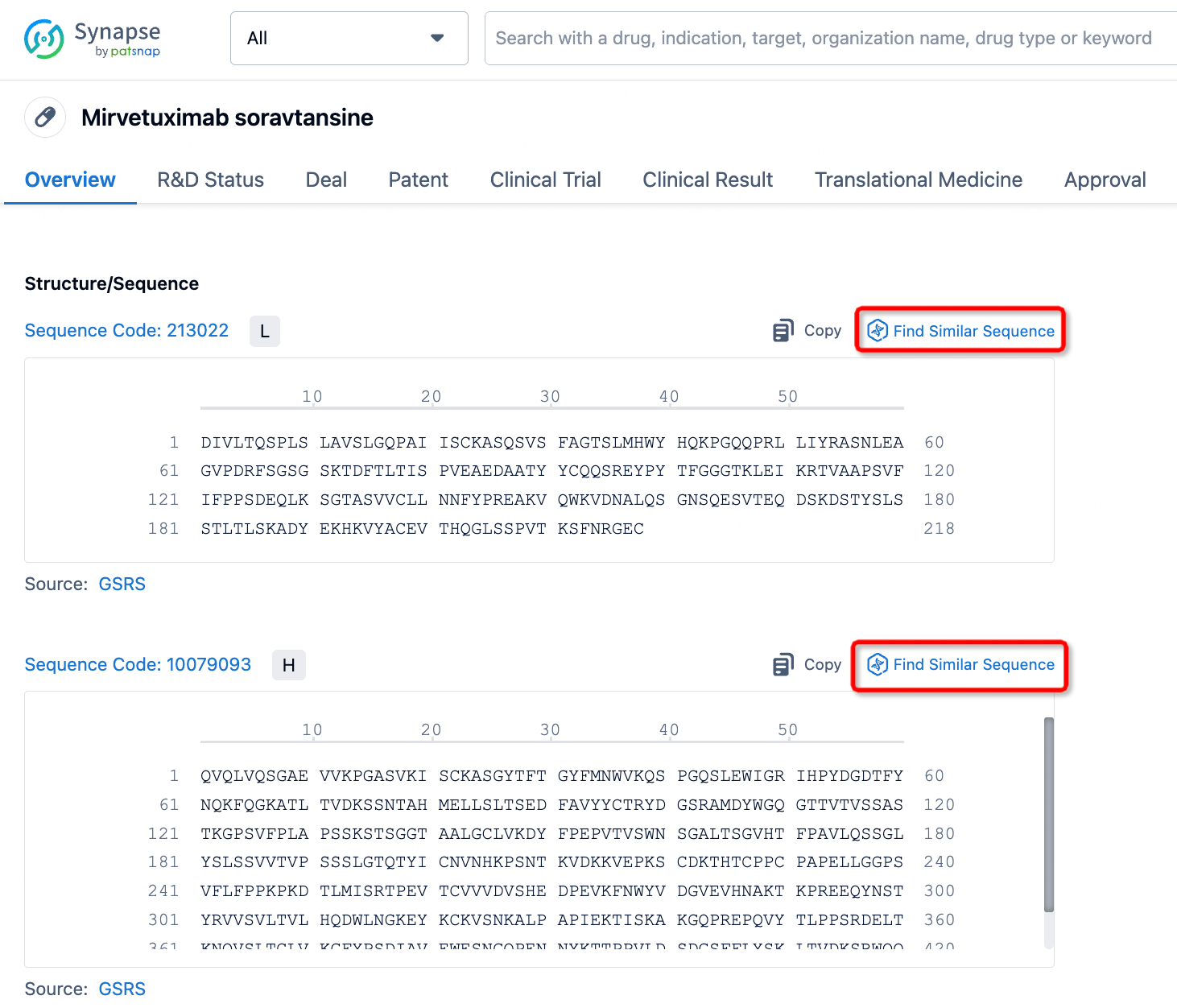
For researchers focusing on the detailed antibody data of a specific ADC drug, Patsnap Synapse offers a direct link to Patsnap Bio, which integrates rich biological data such as protein structures, gene sequences, and biological activity information. Patsnap Bio further contains data on biomarkers, gene expression, and biological pathways, all of which can help optimize ADC design to ensure efficient drug delivery to target cells with minimal side effects.
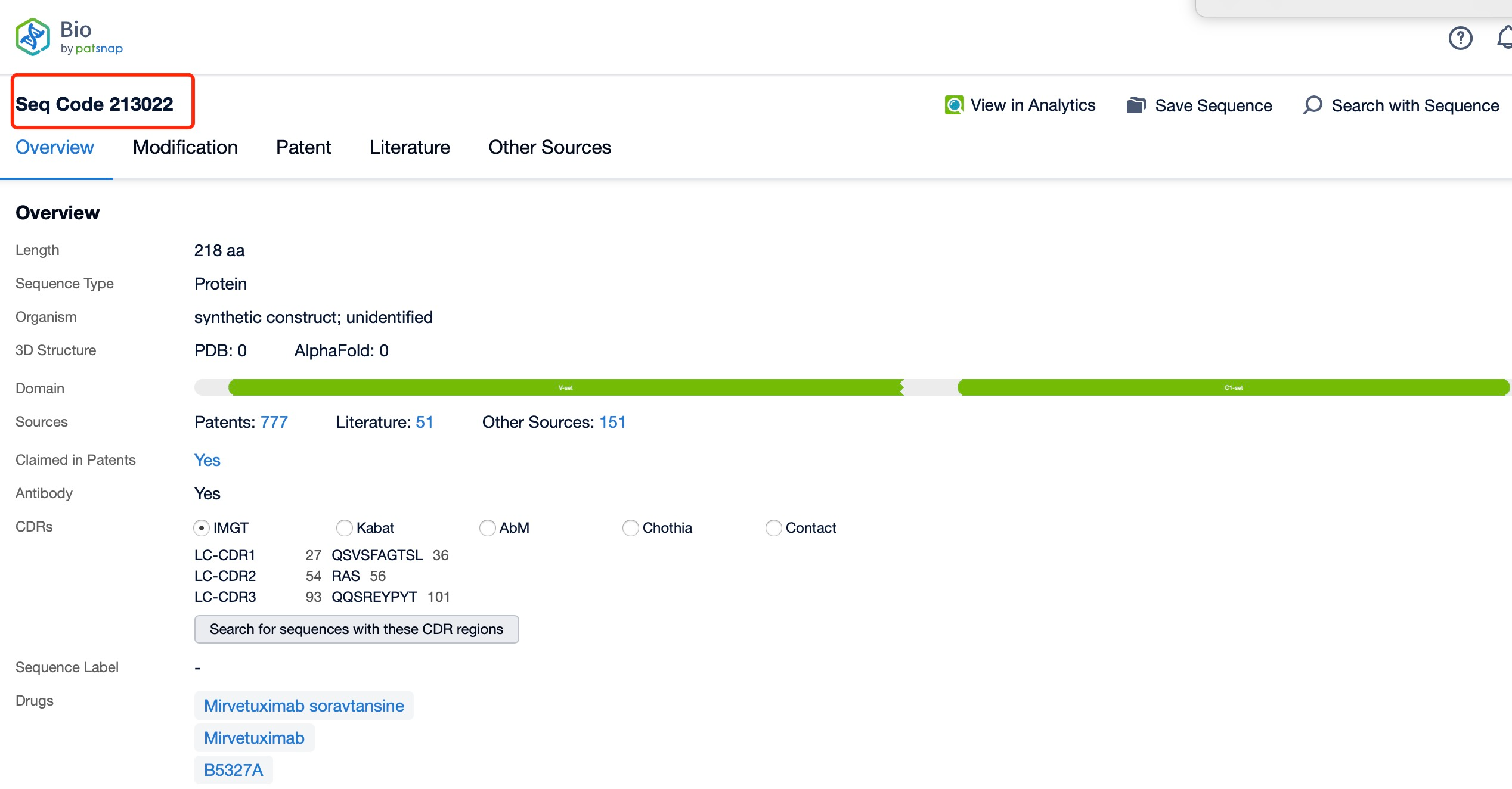
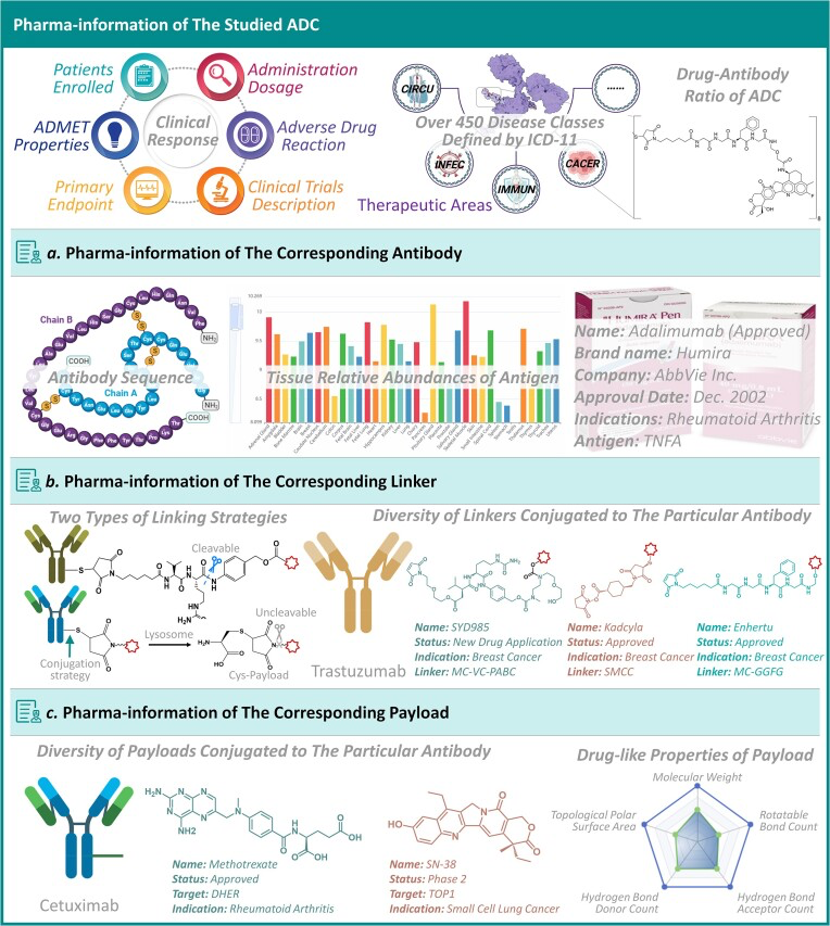
At the same time, Patsnap Synapse links to Patsnap Chemical, allowing researchers to perform structural searches and advanced filtering for ADC linkers. This feature enables easy identification and evaluation of potential linker candidates, including acid-sensitive, protease-sensitive, or disulfide bond-based linkers.
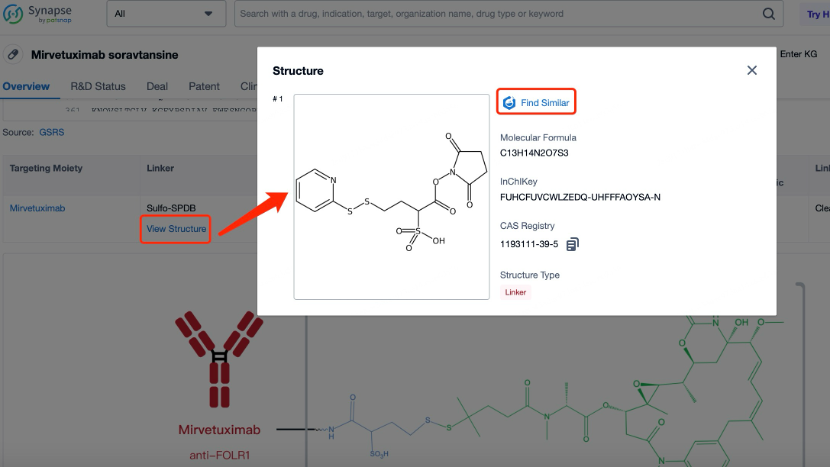
Linker Screening with Patsnap Chemical
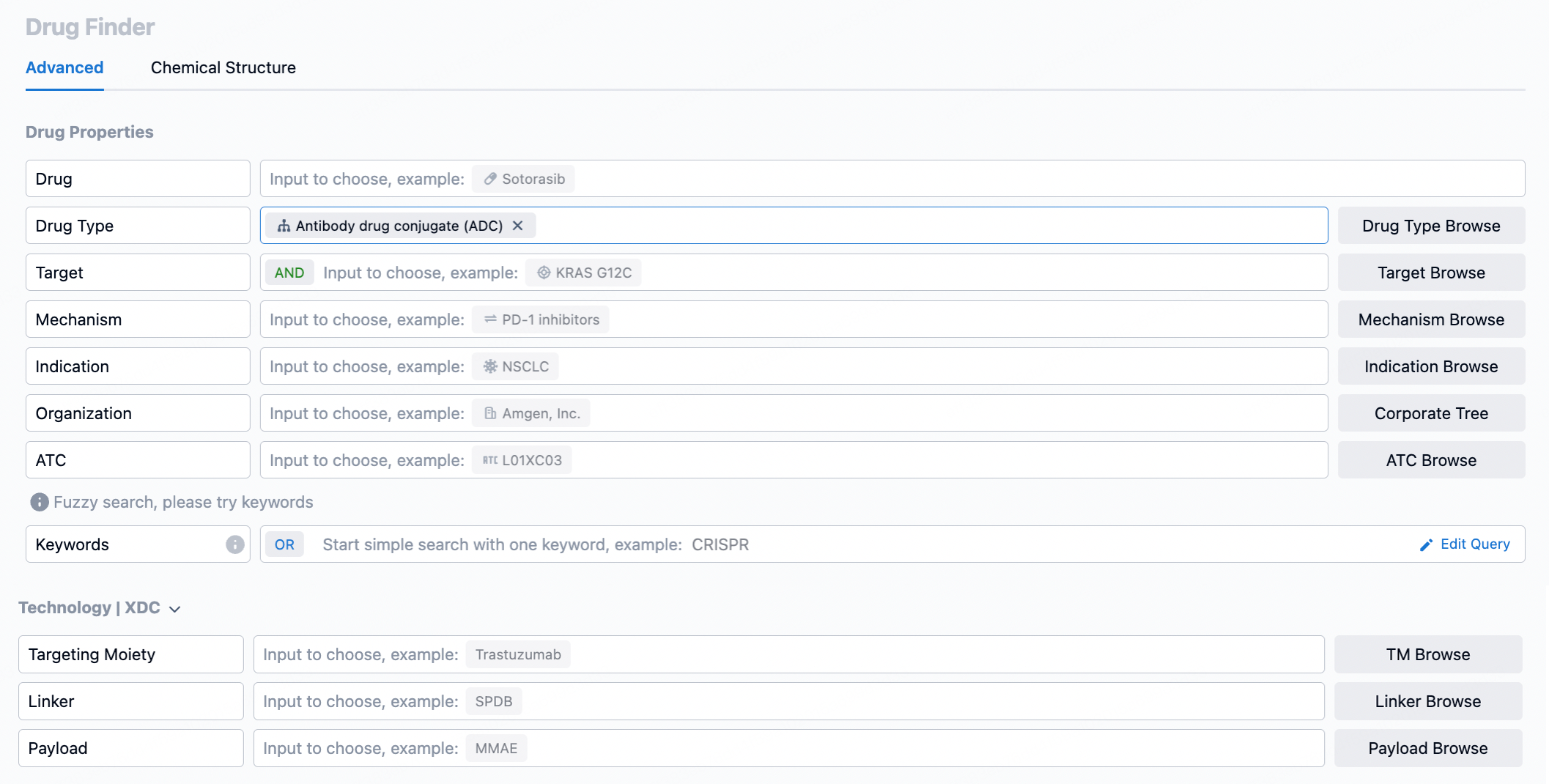
In Patsnap Chemical, researchers can begin their search for ADC linkers using various search methods. Structural search is particularly effective for molecules with specific chemical structures like linkers. By sketching the basic chemical structure, such as one containing specific functional groups, researchers can quickly locate their target compounds.
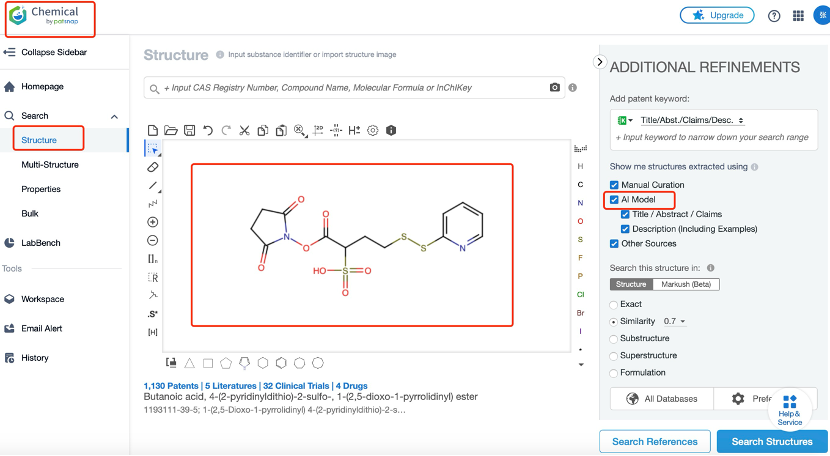
After inputting basic search criteria, the next step involves refining the results through advanced search options. This can be done by setting parameters for the compound's physicochemical properties (e.g., molecular weight, solubility) or biological activity data. For ADC linkers, the focus is on their stability in vivo and controlled cleavage at the target site. Therefore, the filtering conditions should account for linker stability in plasma and cleavage characteristics in the tumor microenvironment to ensure effective drug release upon reaching the target.
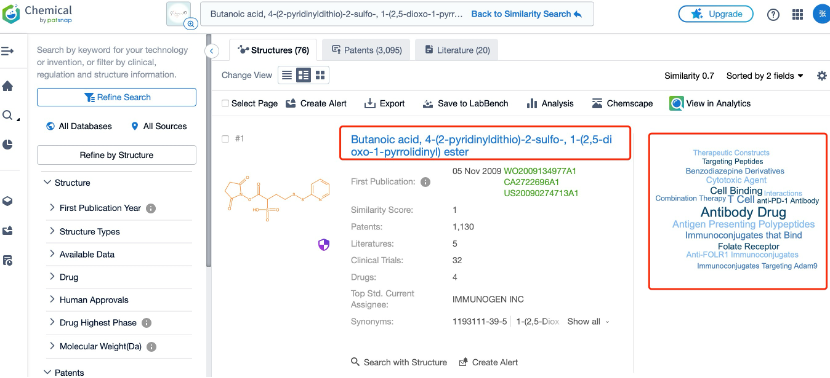
Finally, detailed evaluation of the filtered linkers is essential. This includes reviewing each linker’s chemical structure, synthesis pathway, potential patent status, and relevant literature. Researchers should also assess whether these linkers have been successfully used in existing ADC designs and their performance in preclinical or clinical trials. This thorough evaluation process helps to determine which linkers are most suitable for a specific ADCs development project.
In-depth Exploration of ADCs with Hiro LS
The development of ADCs involves several complex stages, including drug design, preclinical studies, and clinical trials, each with its own set of challenges and bottlenecks. The large-scale biopharmaceutical model Hiro-LS offers solutions and insights across various areas of drug development and market research, enhancing the efficiency and success rate of drug discovery. It not only improves the safety and efficacy of drugs but also provides robust support for the future of personalized medicine.

For an experience with the large-scale biopharmaceutical model Hiro-LS, please click here for a quick and free trial of its features!
Summary
Through machine learning, researchers can rapidly identify linker candidates with optimal chemical and biological stability, significantly reducing screening time. These models are trained on known linkers and their properties, automatically detecting stable molecules and simulating their performance under different physiological conditions, ensuring their stability in practical applications.
The application of AI in ADC development, especially in linker design, greatly enhances efficiency and outcomes. By analyzing vast structural and bioactivity data, AI identifies linker properties that maintain ADC stability in blood circulation while enabling the controlled release of cytotoxic drugs in tumor microenvironments. AI also helps in designing the chemical properties of linkers, such as hydrophilicity or hydrophobicity balance, improving pharmacokinetic properties and ensuring optimal drug distribution and elimination in the body. As AI technology continues to evolve, future ADC development will increasingly focus on personalized and precise treatments. Using advanced algorithms such as deep learning, AI not only refines linker design but also enhances the overall therapeutic efficacy and safety of ADCs.
Reference
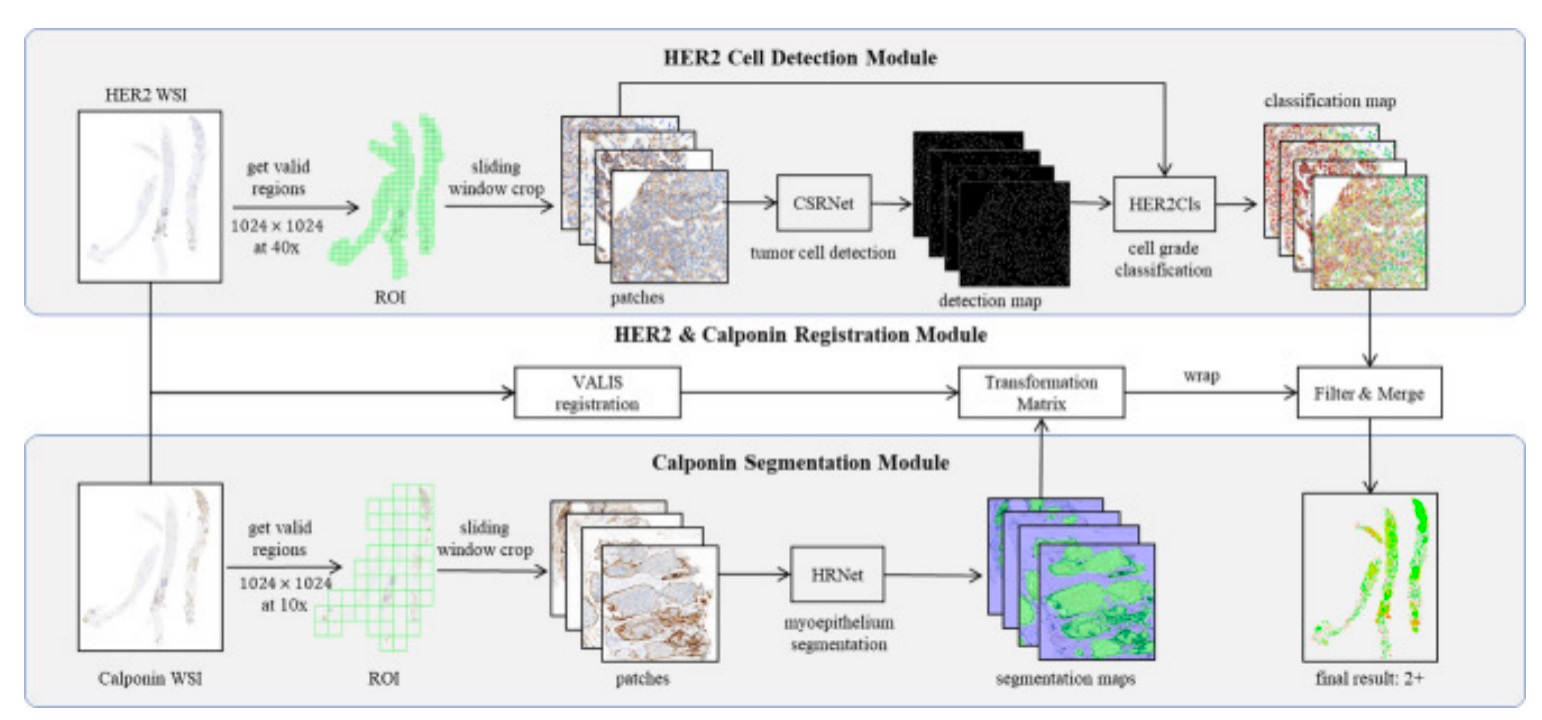
- 1.Xiong, Z., et al., Precision HER2: a comprehensive AI system for accurate and consistent evaluation of HER2 expression in invasive breast Cancer. BMC Cancer, 2024. 24(1): p. 1204.
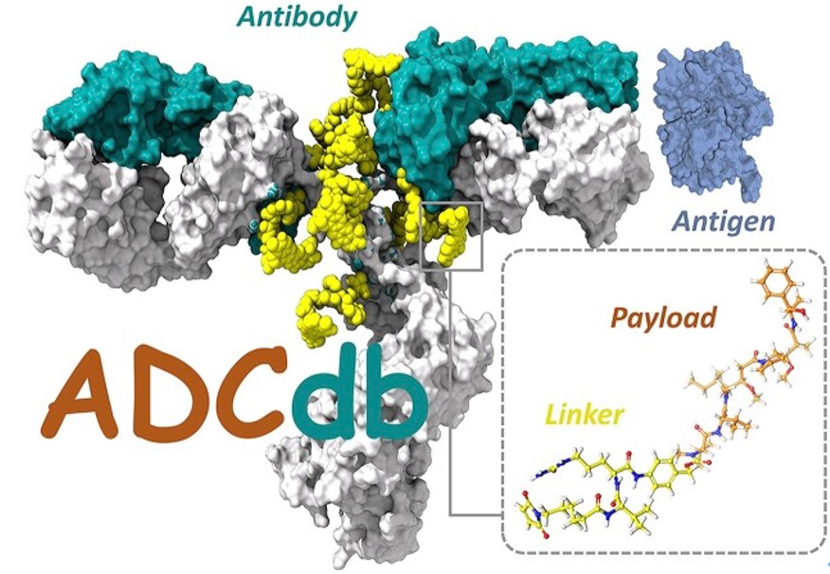
- 2.Shen, L., et al., ADCdb: the database of antibody-drug conjugates. Nucleic Acids Res, 2024. 52(D1): p. D1097-D1109.