How to find the core components of Inotuzumab Ozogamicin?
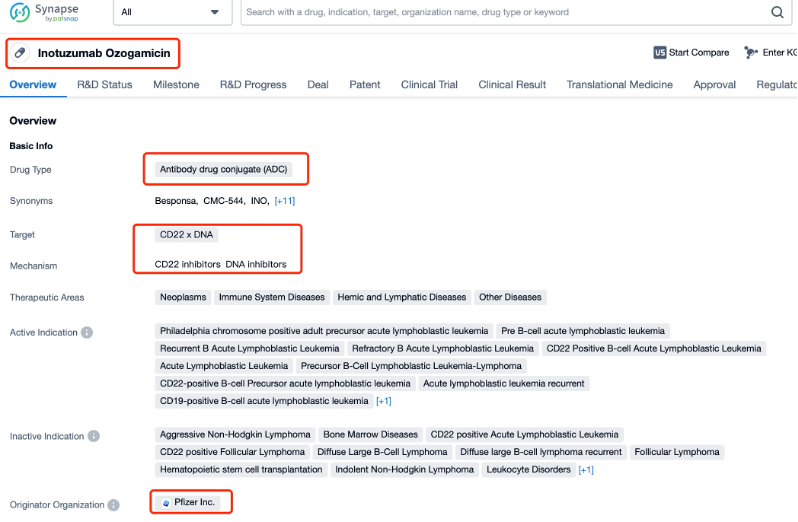
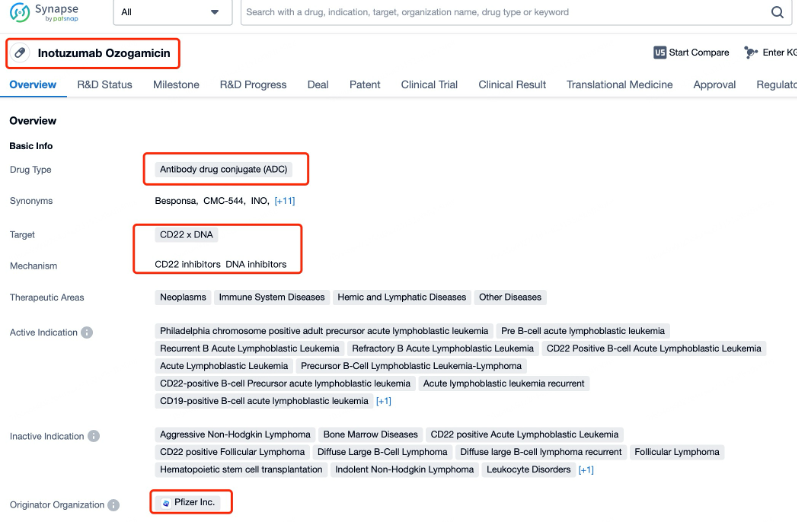
Inotuzumab Ozogamicin, also known as Besponsa, is an antibody-drug conjugate (ADC) developed by Pfizer. It targets CD22, a protein that is expressed on the surface of B cells, including those found in various hematological malignancies such as acute lymphoblastic leukemia (ALL). As a novel therapeutic agent, Inotuzumab Ozogamicin is designed to deliver a potent cytotoxic payload specifically to tumor cells expressing CD22, thereby minimizing toxicity to healthy tissues.
Summary of Research Progress of Inotuzumab Ozogamicin
The research progress of Inotuzumab Ozogamicin has been significant. The drug works by binding to CD22 on the surface of cancer cells, internalizing the ADC, and releasing the cytotoxic payload within the cell. This mechanism allows for targeted delivery of the drug, enhancing its efficacy while reducing systemic side effects. Inotuzumab Ozogamicin has received approval from the U.S. Food and Drug Administration (FDA) for the treatment of adults with relapsed or refractory B-cell precursor ALL. It is approved for use as a single agent. Globally, the drug is also approved in several other countries, with ongoing efforts to secure approvals in additional markets.
The global competition in the ADC market is intense, with several other ADCs targeting various cancers at different stages of development. Key competitors include Genentech's Kadcyla (T-DM1) for HER2-positive breast cancer, Seattle Genetics' Adcetris (brentuximab vedotin) for Hodgkin lymphoma and systemic anaplastic large cell lymphoma, and Astellas Pharma and Seattle Genetics' Enfortumab Vedotin for urothelial cancer. Despite this competition, Inotuzumab Ozogamicin stands out due to its specific targeting of CD22 and its potential in treating ALL, an area with limited treatment options. Clinical trials have shown promising results, particularly in patients with relapsed or refractory B-cell precursor ALL who have progressed on or after multiple lines of therapy. The INO-VATE ALL trial, a Phase III study, demonstrated that Inotuzumab Ozogamicin significantly improved complete response rates and overall survival compared to standard chemotherapy.
Structural Characteristics of Inotuzumab Ozogamicin
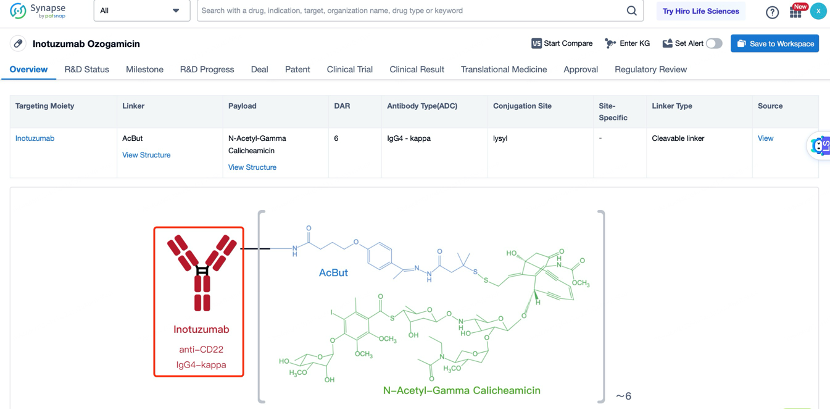
The overall structural characteristics of Inotuzumab Ozogamicin are designed to optimize its therapeutic potential. The ADC consists of three main components: the antibody, the linker, and the cytotoxic payload. The antibody is a humanized IgG4 monoclonal antibody that binds to CD22 with high affinity and specificity. The linker is a non-cleavable calicheamicin derivative linker that ensures the payload remains attached to the antibody during circulation and is released only upon internalization into the target cell. The cytotoxic payload is calicheamicin, a potent DNA-damaging agent.
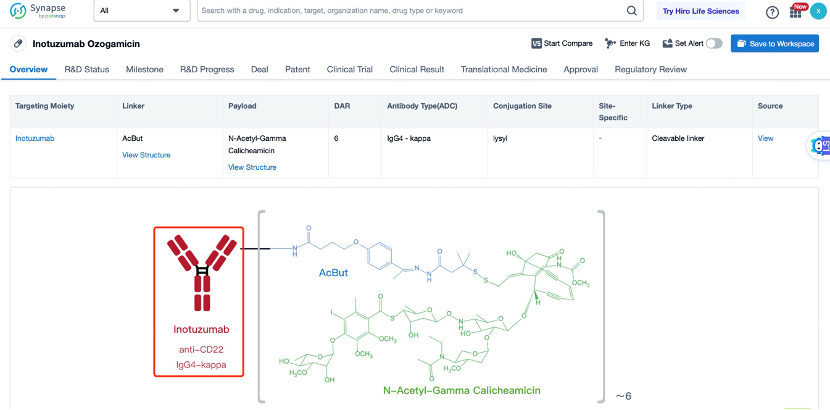
The selection and advantages of the antibody in Inotuzumab Ozogamicin are crucial for its effectiveness. The antibody used, known as huM195, is a humanized IgG4 monoclonal antibody that binds to CD22 with high affinity and specificity. CD22 is a cell surface receptor that is overexpressed in B-cell malignancies, including ALL. The high affinity and specificity of huM195 ensure that the ADC can effectively target and bind to CD22-expressing cancer cells, thereby maximizing the delivery of the cytotoxic payload. Additionally, the antibody has been engineered to enhance its stability and reduce immunogenicity, making it suitable for repeated dosing. The high binding affinity and low immunogenicity of the antibody contribute to the overall safety and efficacy of Inotuzumab Ozogamicin. The antibody also has a favorable pharmacokinetic profile, with a long half-life that allows for less frequent dosing, improving patient convenience and compliance.
The linker in Inotuzumab Ozogamicin is a key component that ensures the stability of the ADC in circulation and the controlled release of the payload inside the target cell. The linker used is a non-cleavable calicheamicin derivative linker. This linker is stable in the bloodstream and remains intact during circulation, ensuring that the payload does not prematurely detach and cause systemic toxicity. Once the ADC is internalized into the target cell, the payload is released through the action of lysosomal enzymes. This design minimizes the risk of off-target toxicity and ensures that the cytotoxic agent is delivered directly to the tumor cells. The non-cleavable nature of the linker also provides a high degree of control over the release of the payload, enhancing the therapeutic index of the ADC. The linker is designed to maintain the integrity of the ADC during systemic circulation, preventing the payload from being released before reaching the target cells. This stability is crucial for minimizing systemic toxicity and ensuring that the drug reaches its intended target. The non-cleavable linker also provides a high drug-to-antibody ratio (DAR), typically around 2, which enhances the therapeutic index of the ADC.
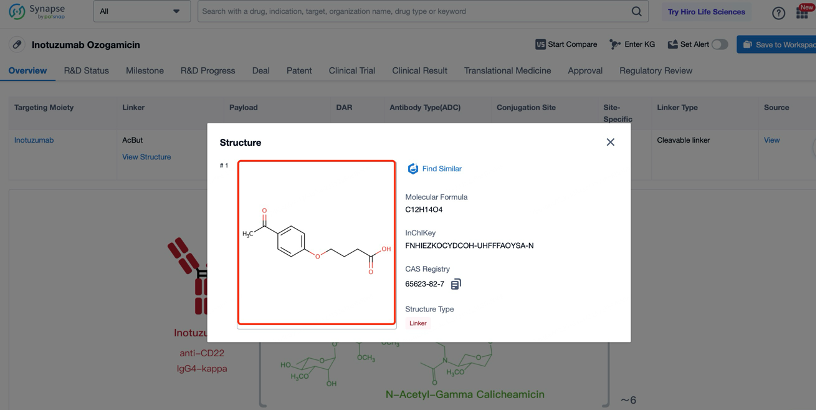
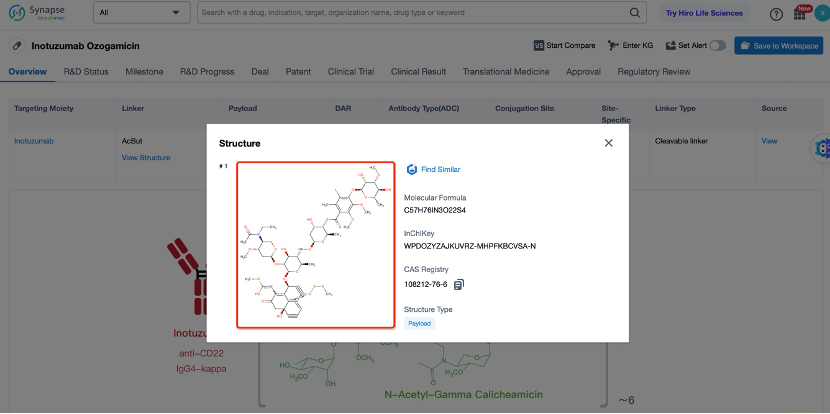
The cytotoxic drug payload in Inotuzumab Ozogamicin is calicheamicin, a potent DNA-damaging agent. Calicheamicin works by forming a covalent bond with the sugar-phosphate backbone of DNA, leading to double-strand breaks and cell death. Calicheamicin is chosen for its high potency and ability to induce cell death at low concentrations. The payload is linked to the antibody through the non-cleavable linker, ensuring that it remains inactive during circulation and is only activated once inside the target cell. This design enhances the safety and efficacy of the ADC by minimizing systemic toxicity. Calicheamicin is particularly effective against cancer cells because it targets DNA, which is essential for cell division and survival. By causing double-strand breaks in DNA, calicheamicin prevents cancer cells from completing the cell cycle, leading to cell death. The high potency of calicheamicin allows for the use of lower doses of the ADC, further reducing the risk of side effects. Additionally, calicheamicin has a broad spectrum of activity against various cancer types, making it a versatile choice for the development of ADCs.
Summary and Prospect
In summary, Inotuzumab Ozogamicin represents a significant advancement in the treatment of acute lymphoblastic leukemia (ALL), particularly for patients with relapsed or refractory B-cell precursor ALL who have progressed on or after multiple lines of therapy. The drug's unique mechanism of action, combined with its optimized antibody, linker, and cytotoxic payload, positions it as a promising therapeutic option. Ongoing clinical trials continue to evaluate its safety and efficacy, and if successful, Inotuzumab Ozogamicin has the potential to become a standard treatment for ALL, addressing a critical unmet medical need. Future research will focus on expanding its use to other CD22-expressing cancers and exploring combination therapies to further enhance its therapeutic benefits. The detailed selection and engineering of the antibody, the stability and non-cleavable nature of the linker, and the high potency of the cytotoxic payload all contribute to the overall effectiveness and safety of Inotuzumab Ozogamicin, making it a promising candidate in the field of targeted cancer therapy.
How to find the the core components of an ADC drug?
In Patsnap Synapse and Patsnap Bio, you can find the sequence and latest research and development advances of all ADC drugs.
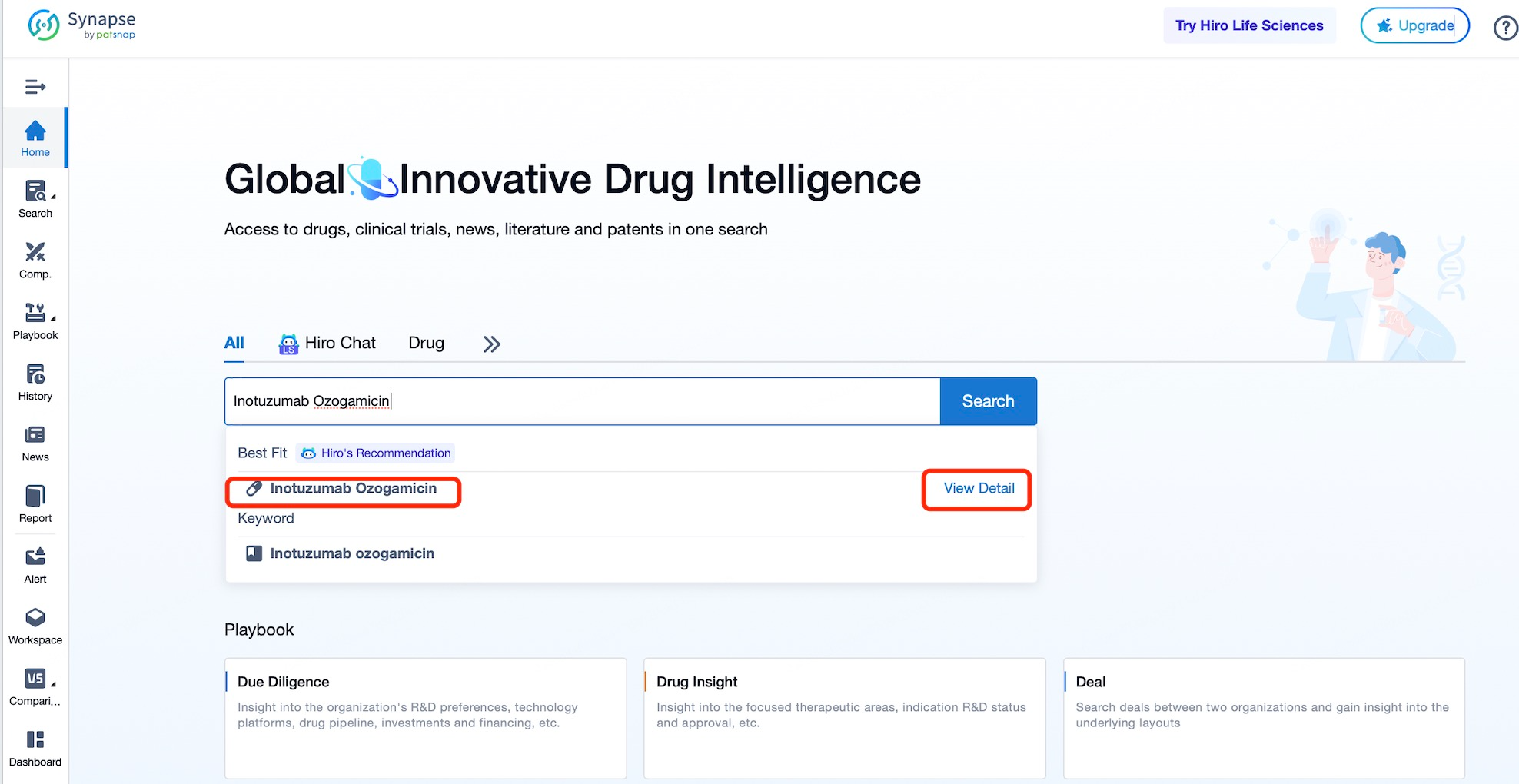
Taking Inotuzumab Ozogamicin as an example, First, you can log in to Patsnap Synapse, enter ' Inotuzumab Ozogamicin ' in the search box and click to view the details. On the details page, you can find the basic information and research progress of Inotuzumab Ozogamicin.
After entering the details page, drop down to find the core Structure information of ADC drug and click view Structure in the Linker section to find the structure and type of ADC drug Linker.
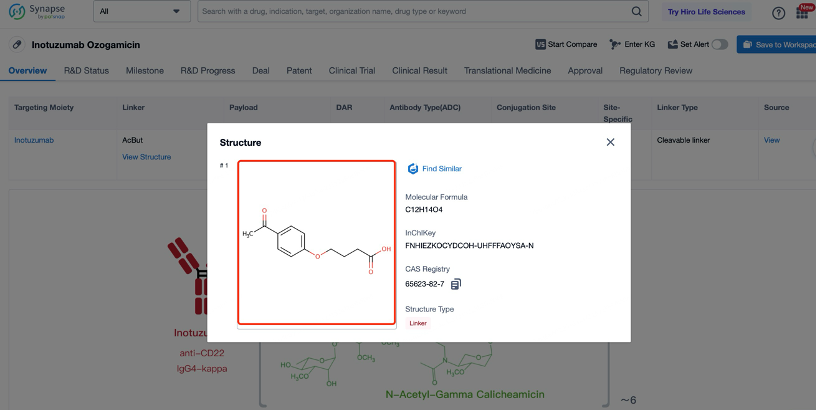
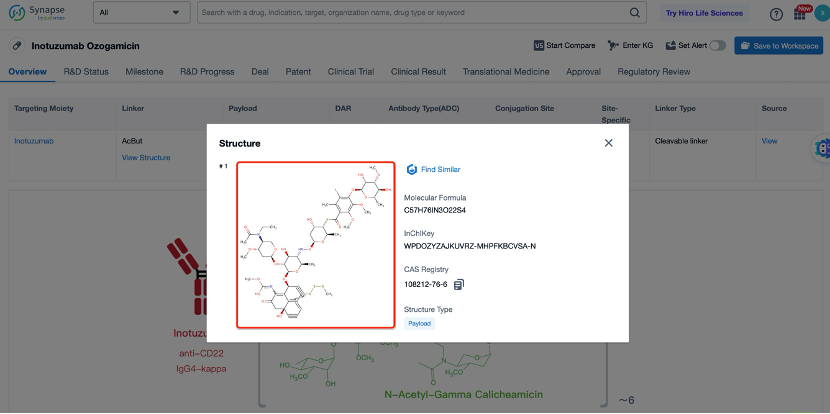
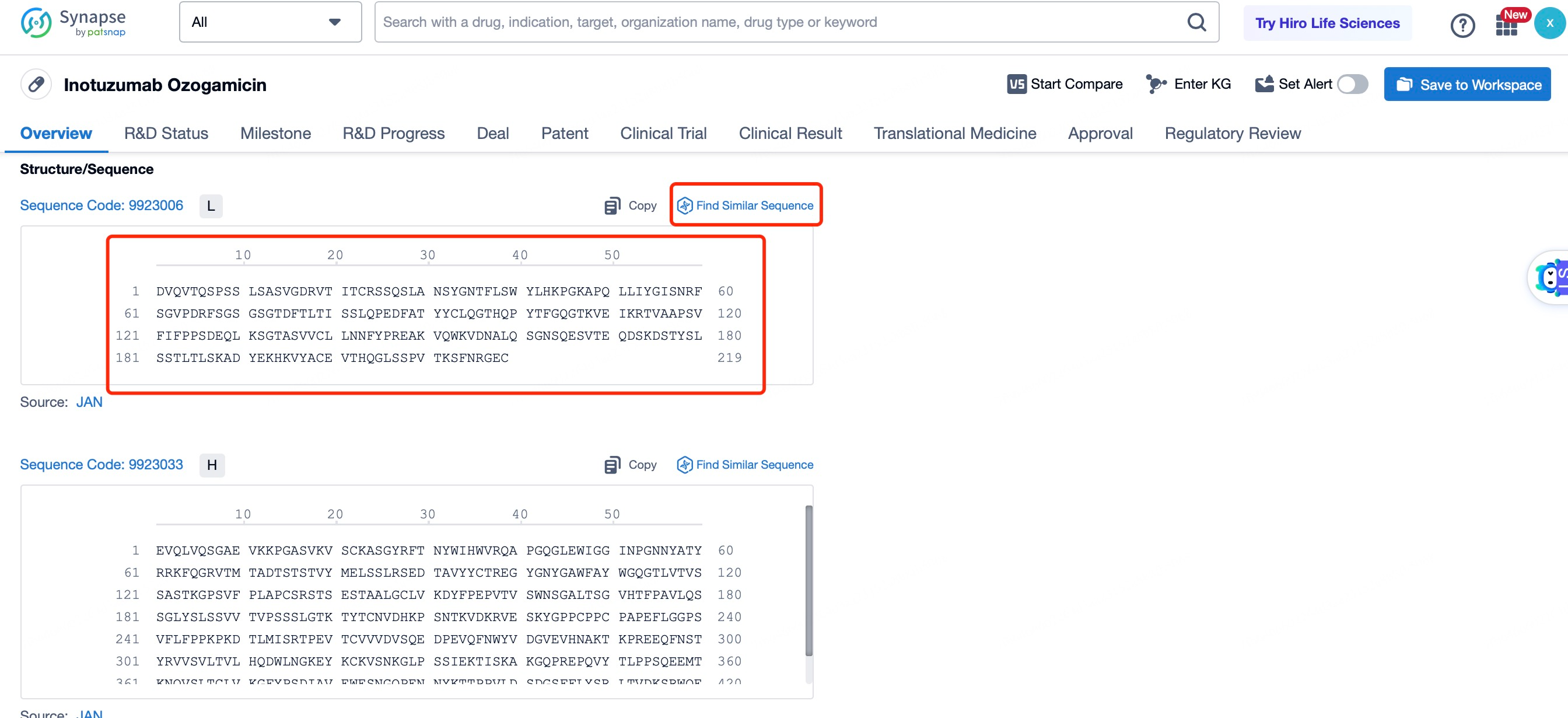
The details page also lists the complete sequence information of the antibody part. By clicking on "Find Similar Sequence", you can be redirected to Patsnap Bio to search for similar sequences of the antibody.
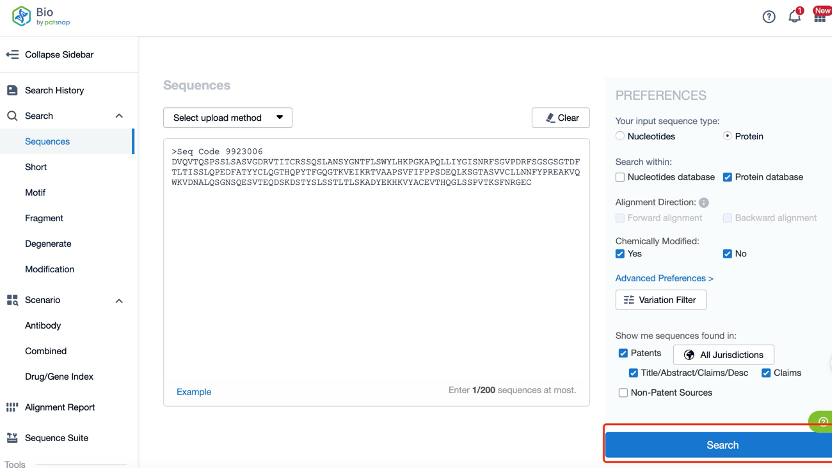
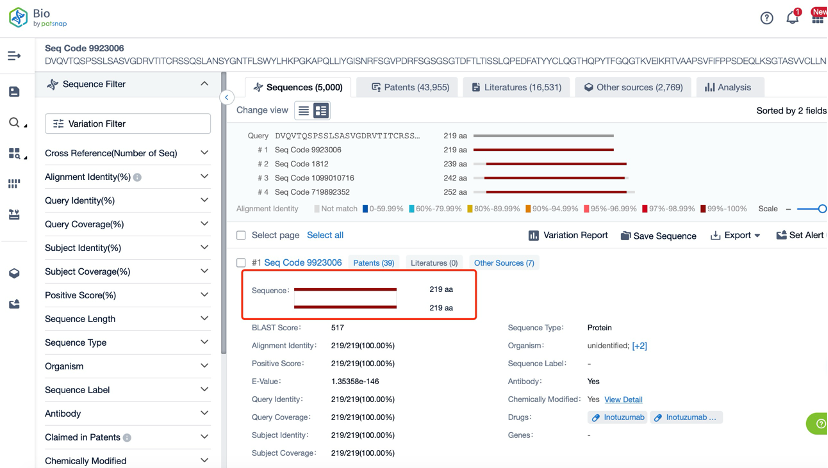
Clicking on the sequence name will provide you with all the basic information of that sequence.
Patsnap Bio helps you turn weeks into minutes with cutting-edge AI-enabled tools built to master the complexities of sequence retrieval and automate IP analysis with precision and ease.